Eukaryotic Transcription Regulation
Vocabulary:
Cell-type specific expression: Different cells may express very different sets of genes, despite containing the same DNA, allowed for by gene regulation
Chromatin: While DNA is in this structure, transcription, replication, and DNA repair are all inhibited
Nucleosome: The fundamental subunit on chromatin, 2 turns of DNA wrapped around 8 histones, making an octamer
Histone: A protein that DNA wraps around, small and positively charged, largely Arg and Lys, bind to DNA and are highly conserved
Histone H1: The protein that binds to linker DNA between nucleosomes forming the macromolecular structure known as the chromatin fiber
Chromosome territory: Within a nucleus in interphase, chromosomes will occupy discrete domains and stay separate from other chromosomes
Interchromosomal domain: Channel between chromosomes that contain little or no DNA
Transcription factories: A feature within the nucleus that may contribute to gene expression, contain most active RNAP and transcription regulatory molecules, they are dynamic structures that form rapidly and disassemble upon stimulation and repression of transcription. They are at the edge of chromosome territories and move into the adjacent interchromatin compartments. They house RNA processing machinery and are contiguous with nuclear pores.
DNase I hypersensitivity: Results in more open chromatin configuration states, upstream of the transcription start site
Histone modification: The addition of acetyl groups to histone proteins and histone protein tails, and the addition of methyl groups to histone protein tails and DNA
“Closed” chromatin: An inhibitory conformation of DNA, where DNA is tightly associated with histones
DNase I: Digests DNA to varying degrees depending on the conformation
H2A.Z: Affects nucleosome mobility and positioning on DNA, found more at promoters and enhancers
Histone modification: Involves covalent bonding of functional groups onto N-terminal tails of histones
Histone acetyl transferase enzymes (HATs): Associated with increased transcription, catalyze the addition of an acetyl group to a histone to make the charge more positive and cause the histone to bind less strongly do DNA
Histone deacetylase (HDAC): Remove acetyl groups from histone tails, associated with a decrease in transcription
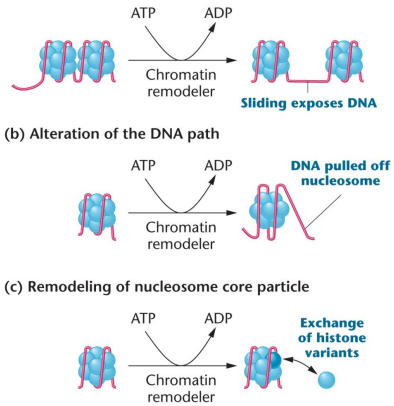
Chromosome remodeling: Involves the repositioning or removal of nucleosomes on DNA, multi-subunit enzymes use the energy of ATP hydrolysis to remove and arrange nucleosomes. Directed by binding to transcription factors, modified histones, or methylated DNA
DNA methylation: Enzyme mediated addition or removal of a methyl group to DNA, done by a methyltransferase, occurs most commonly at position 5 of cytosine, causing it to protrude into the major groove of the DNA helix, can decrease gene expression, tissue specific
CpG Island: A region with many methyl groups on DNA, occurring near the promoter
Silenced DNA: Happens when there are many methyl groups attached to DNA, this is what happens to the Barr body
Cis-acting sequence: Located on the same chromosome as the gene it regulates, required for accurately regulated transcription on genes, either a promoter, enhancer, or silencer
Promoter: A nucleotide sequence that serves as a recognition site for transcription machinery, adjacent to regulatory genes and critical for transcription initiation
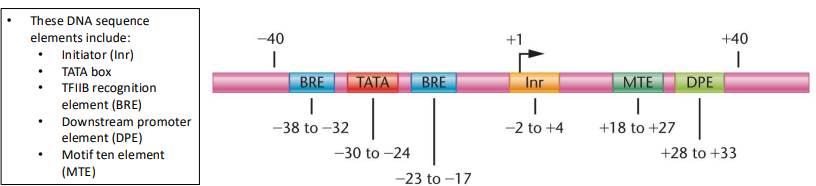
Core protomer: A protomer that determines accurate initiation of transcription, minimum required for accurate initiation of RNAP II, around 80 nt long, has the transcription start site (TSS)
Proximal promoter elements: Modulate efficiency of transcription, around 250 nts upstream of the TSS, have binding sites for sequence specific factors that modulate the efficiency of transcription
Focused promoters: Specific transcription initiation at start sites, major type of initiation for lower eukaryotes
Dispersed promoters: Direct initiation from several weak transcriptional start sites located over 50-100 nt region
Enhancers: Located on either side of a gene, or within, important for reaching max levels of transcription, cis elements, orientation/position independent, confer time/tissue specific gene expression, modular and have many binding sequences that additively enhance effects
Green Fluorescent Protein (GFP): Emits a green fluorescence when excited by blue light, used in reporter genes, can show effect of an enhancer on transcription
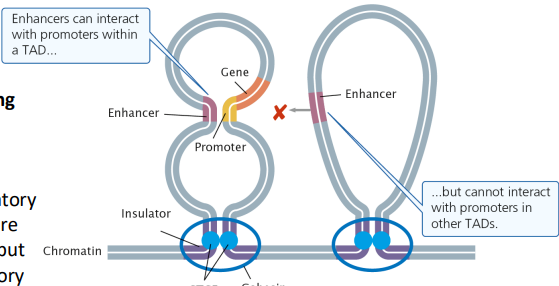
Insulators: Found between the enhancer and promoter for a nontarget gene, limits the extent of enhancers, enable and block enhancer/promoter interactions
Silencer: Can be on either side of a gene, or within, acts as a negative region of transcription, represses levels of initiation, act as a tissue/time specific way to control gene expression, binding site for repressor proteins
Transcription factors (Tx): Regulatory proteins involved in transcription, target cis-acting sites
Activator: A kind of Tx that increased transcription initiation
Repressor: A kind of Tx that decreases transcription initiation
Human metallothionein IIA gene (hMTIIA): A protein that binds to heavy metals and protects cells from toxic effects of high levels of them, protects cells from oxidative stress, can be upregulated
DNA-binding domain: Part of a Tx, binds to specific DNA sequences in the cis site, forms hydrogen bonds with DNA, fits into the major groove
Trans-repressing domain: Part of a Tx, activates/repressed transcription by binding to other transcription factors or RNAP
Helix-turn-helix (HTH): Part of a DNA-binding protein, present in both eukaryotic and prokaryotic Tx, monomer has 2 alpha helices, binds to the major groove
Zinc finger: Part of DNA-binding proteins, uses Zn to stabilize structure, has many protrusions that make contact with the target molecule
Basic leucine zipper (bZIP): Part of DNA-binding proteins, allows for protein-protein dimerization, has a region that mediates sequence specific DNA properties, holds together 2 DNA binding regions
Pre-initiation complex: Formed by the assembly of proteins in a specific order, provides a platform for RNAP II to recognize the TSS. Formed by TFIID binds to the TATA box via TBP to start forming this, followed by the addition of TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, and the final mediator
Coactivator: Interacts with proteins and enables an activator, form an enhanceosome
Enhanceosome: Formed by coactivators, interacts with the transcription complex
GAL gene system: 4 structural and 3 regulatory genes, products transport galactose into the cell to metabolize it, inducible, genes not transcribed without galactose and positively regulated by galactose
GAL4: The regulatory gene that activates proteins that regulate GAL structural product transcription
GAL4 Protein (GAL4p): Encoded by GAL4, negatively regulated by Gal80p, has a DNA binding domain that recognizes and binds to sequences in UAS
ENCODE: The encyclopedia of DNA elements project, launched in 2003 to identify all functional DNA sequences and determine how elements regulate expression in humans, determined less than 2% of the genome is proteins, with over 80% being regulatory of these
Enhancer elements: Contain promoter elements that initiate transcription
Enhancer RNA: A genomic sequence that encodes enhancer RNA, often contain promoter elements that initiate transcription from the enhancer and extend outward for several kilobases
Genomic wide association study (GWAS): Identifies genetic variations that are significantly enriched in the genomes or patients with a particular disease and are not found in those without the disease, can evaluate single base pair changes or SNPs
With eukaryotes, regulation can occur during chromatin remodeling, splicing, nuclear export, mRNA stability, mRNA localization, translocation, and protein modification. Genes are on chromosomes, and are combined with histones
Within a chromosome, structure is continuously rearranging, transcriptionally active genes are cycled to the edges of a chromosome’s territory
SWI/SNF binding will cause the nucleosome to arrange in a way where DNA histone contacts may be loosened (allowing the nucleosome to slide along the DNA and expose regulatory regions), the path of DNA around a nucleosome core particle may be altered, or components of the core nucleosome particle may be rearranged such as swapping in and out variable histone proteins
A protein will bind to an insulator and induce the formation of a DNA loop, which will allow specific enhancer-promoter interactions and not others
Galactose metabolism is inducible, transcription is activated by GAL4 in response to galactose, where GAL4p binds to the UAS site and controls transcription levels, UAS has the properties of an enhancer, as it is a regulatory sequence lying upstream of the gene.