lecture 7: secondary transporters
introduction
secondary active transporters use chemical gradients to drive transport. There are different types of transporters such as uniporters that bind one molecule that goes across the membrane down a concentration gradient. Symporters co transport with ion and the substrate to allow the substrate to be transporters up the concentration gradient. Antiporters take up two substrates in opposite directions. Symporters and antiporters use active transport. substrate here denotes any molecule being taken up as transporters can be seen as physical enzymes
transporters use an alternating access mechanism. The transporter by itself can switch easily between inward and outward facing states. For uniporters, one molecule binds and it is flipped across the membrane. At any point, this is reversible. Symporters can also switch when they are in an unbound state but when one molecule binds (usually the ion), it cannot switch to the inward facing state without the second molecule binding.Antiporters have one ion binding and this changes the conformation when the transporter is inward/outward facing to drive the uptake of the second ion.
This is important both physiologically for:
nutrient uptake
maintenance of pH or salt concentrations
neurotransmission
as well as pharmacologically:
inhibitors used as drugs in various diseases
can pump drugs out of cells
can pump drugs into cells
neurotransmission
neurotransmission is packaged in vesicles which is released when the signal comes. This system has to be reset so the signal can be transmitted again. This is done by transporters which allows molecules to be taken back into the axon. These can be used as drug targets for drugs of abuse as well as depression etc
glucose transport
you can reabsorb glucose from the filtrate and there is a low level of glucose in the filtrate. to drive the reabsorption of glucose, a sodium gradient is used. in the cell there is a high concentration of glucose which go through uniporters down its concentration gradient into the blood. sodium ions also flow into the cell which are pumped out by sodium potassium pumps
families of secondary transporters
secondary transporters are classified into families based on function and sequence. these cluster into superfamilies of which the largest are:
major facilitator superfamily (MFS)
this is the largest superfamily of where there are more than 70 families
aminoacid/polyamine/prganoCation superfamily (APC)
theis contains a large number of diverse families
it is only in the last few years that we have been able to solve the structure of mammalian proteins. much of our initial understanding of secondary transporters comes from bacterial proteins. membrane proteins are unstable out of the membrane and proteins from bacteria are more stable than eukaryotic proteins. bacteria also often have homologues of human proteins. these can give the broad principals of the transport mechanism in their human counterparts
APC superfamily
This contrans transporters such as neutransporter transporters. The APC transporters is also known as LeuT transporters as the leucine transporter was the first one solved in this family. It was solved from its bacterial homologue.
All structures that were first solved were bacterial transporters with their human counterparts solved as a consequence. There can be sodium coupled transporters, proton coupled transports and antiporters in this superfamily.
Mhp1
Mgp1 is a sodium symporter. It has a sodium biding site:
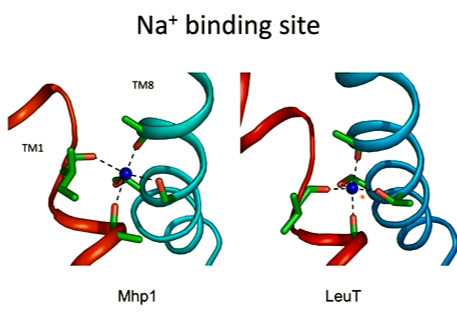
One of the first things noted was the position of the Na ion which was located in a similar position to its LeuT bacterial counterpart. The Na ion here is located at the broken TM1 region which allows the carbonyl oxygen to interact as well as the negative dipole that interacts with the positive ion. The substrate of this interacts at the centre of the protein sandwiched between two tryptophans. The substrate binds tightly at the middle and makes specific interactions
The similarity is only in 10 helices. There is always an inverted V in the structure. Pulling part the 5 N terminal helices and the 5 C terminal helices, the V from both sides are related. the protein is made up of the two units that are known as the inverted repeat. There most likely was an ancestral protein that was made of the first 5 proteins expressed into the dimer. Over time, there was a gene duplication that resulted in the protein we see today.
the alternating access mechanism
the structures were solved of Na in an outward conformation was solved, the empty inward conformation and the sodium and substrate outward conformation was solved. The surface representation was looked at which conformed different conformations:
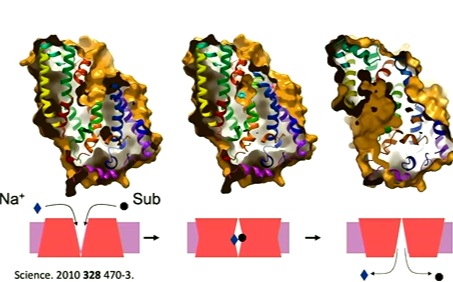
going from unbound to bound is simple as it is only a single helix that moves. The substrate entres and the helix 10 rotates and blocks the substrate in the binding site when it entres. TM10 has become to known as having a thin gate and a thick gate where TM10 is the thin gate. There are more conformational changes going from being outward facing and inward facing. Superimposing the bundles, the inward and outward structures look similar. Taking the superimposition and only looking at the hash motif, show the bundle doesn’t match very well but superimposing based on the hash motif, you see they match up well. the change is a 30 degree rotation around one axis. This is known as rigid body movement where one motif rotates onto the other.
Sodium binding occurs between heices 1 and 8 (1 on the bundle motif and 8 on the hash motif). This is well formed in the outward facing state but in the inward facing state, the binding site is destroyed as there is movement of the hash motif. This is also true to a lesser extent to the ligand binding site. The hash domain here is known as the thick gate as there is more protein moving between conformations.
sodium ions flowing down their concentration gradient drives the transport. the high concentration of them outside and low of the substrate. there is a conformation of protein that changes when the sodium entrees that fixes the transporter in the outer facing conformation. when the substrate bonds it can swap confirmations.
serotonin transporter structure has also been solved and they first did it with crystallography and expressed it i mammalian cells with 3 stabalising mutations. going from the outward facing to inward facing conformations, there is the rigid body movement but there is more TM1 movement. human dopamine transporter structure was also solved.
MFS superfamily
these include glucose transporters. The first classical transporter studied was LacY that transports lactose into bacteria and is a symporter that is driven by a protein gradient. There is a 2 fold axis as TM 1-6 is found on the right side and Tms 7-12 are found on the left. This means that it is now in the lane of the membrane. The substrate binds in the middle
This also works by the alternating access mechanism
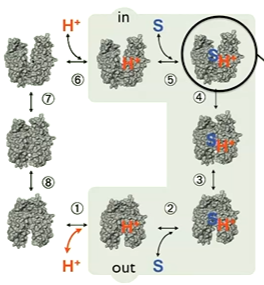
Protons aren’t visible in crystallography so it is important to see which residues are important and are glutamate and arginine. the position of the arginine with affect the pKa of glutamate which affects whether or not the glutamate takes up the protein and so the ligand
glucose transporters ae uniporters and so the binding of glucose depends on the concentration either side of the membrane. This was first discovered by the xylose transporter that has a similar structure with the addition of one domain. The residue protonated here is driven by the proton gradient. Superimposing the structure to the c terminal domain of the xylose transporter show the movement of one of the domains relative to the other.
The crystal structure of the glucose transporters such as Glut5 are known known and these function in a similar way to the xylose transporter. The details are different and so these details must be known when working out drug therapies
MFS transporters have a parallel repeat with this arrangement:

the inverted repeat is a good way and is wide spread through secondary transporters when switching from the inward conformation ot the outward.