6.4 DNA Replication
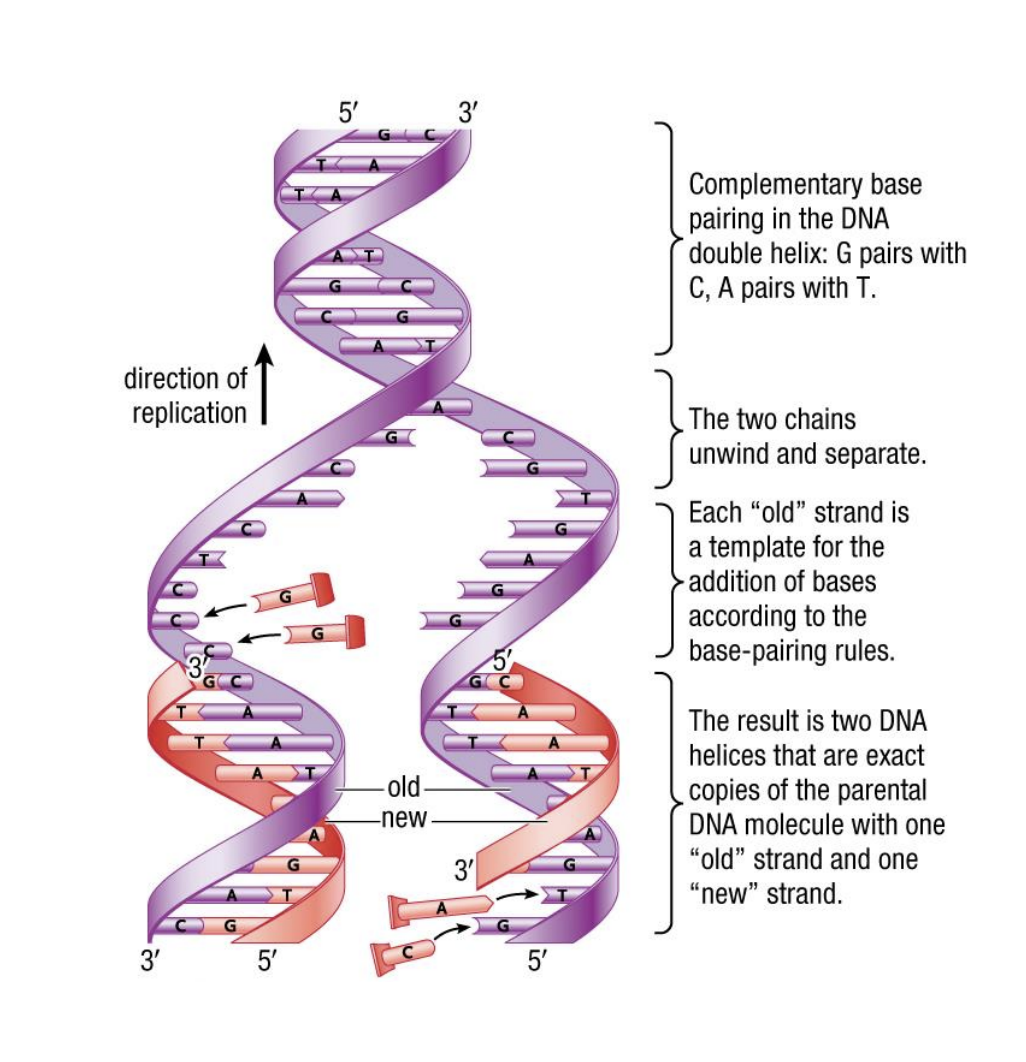
<<Semi Conservative Replication<<
^^Matthew Meselon and Franklin Stahl^^ in 1958
DNA is double stranded
2 parent strands are seperated
complementary strand is made from each parent strand
results in 2 double stranded molecules, each containing one old strand and one new strand
<<Step 1: Separation<<
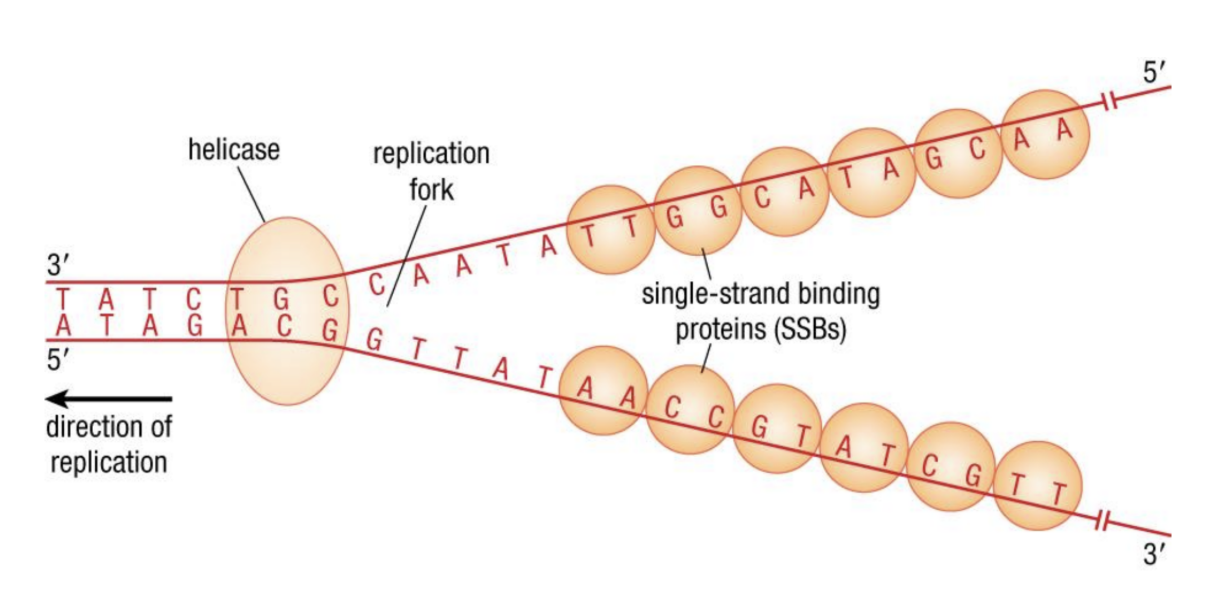
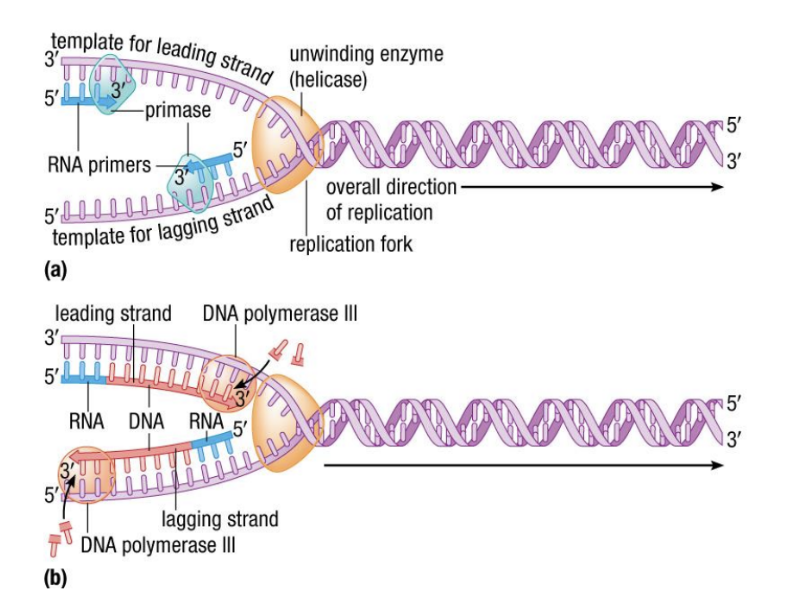
Strands are unwound from ^^replication origins^^ by %%DNA helicase%%==(1)==.
- Forms a ^^replication fork^^
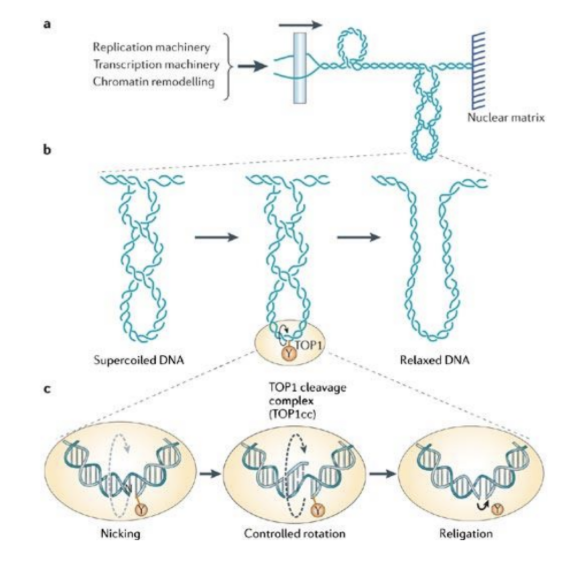
%%Topoisomerases%%==(2)== cut and rejoin strands to keep them from tangling (supercoils)
%%Single-strand binding proteins%%==(3)== attach to unwound strands to prevent them from rejoining (annealing)
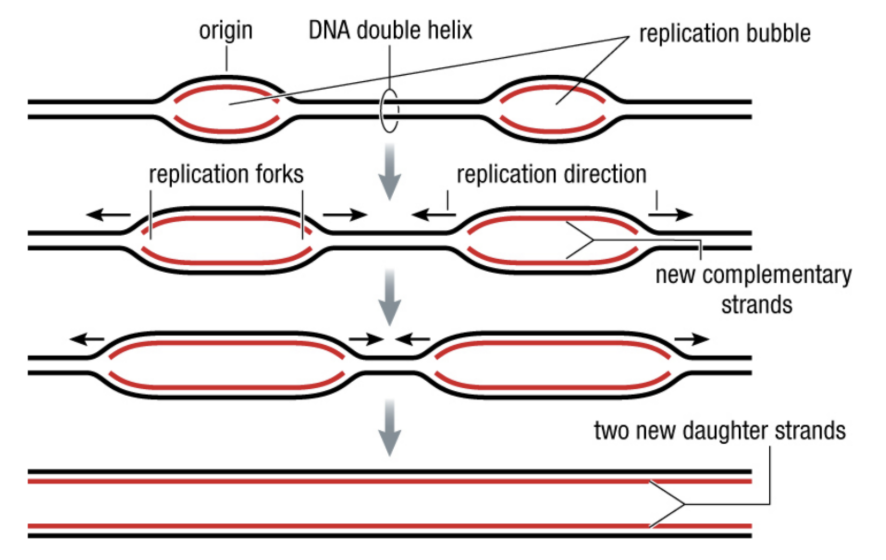
<<Replication Bubbles<<
because there are many replications going on, especially in Eukaryotic DNA
Helicase unwinds DNA in both directions from each origin, producing replication bubbles
Complementary strands are synthesized as the forks continue (step 2)
Bubbles meet and merge, producing 2 separate daughter strands
<<Step 2: Synthesis / Replication<<
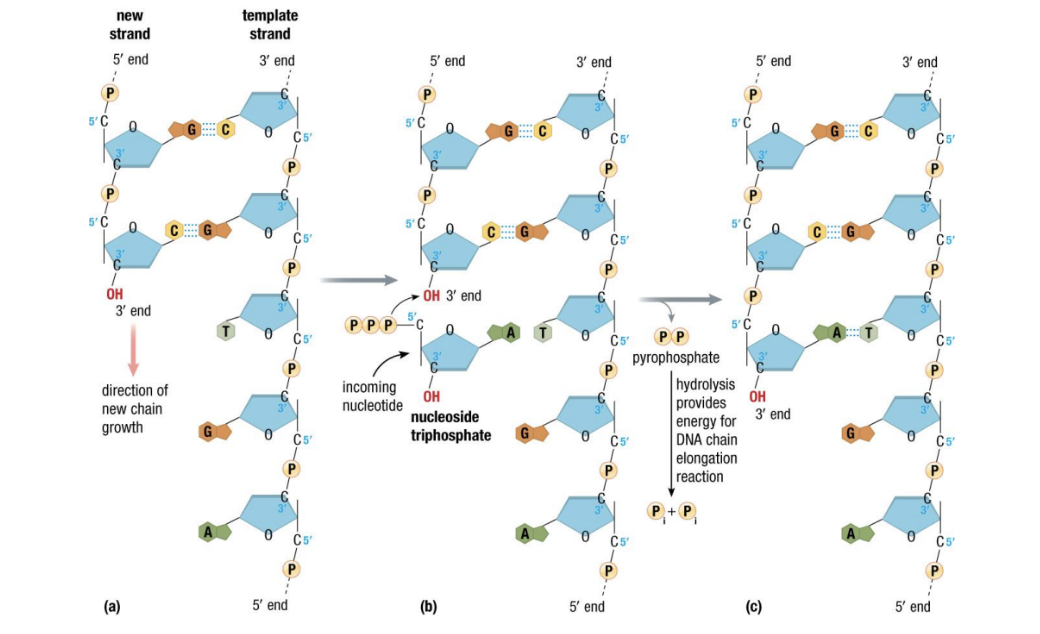
%%Polymerases%% build complementary strands in 5’ → 3’ direction
- can only add to 3’ carbon
- which means its reading 5’
- new strands are always 5’ → 3’
Nucleotides are added as ^^nucleoside triphosphates^^
- a building block and energy source for replicating DNA
- very similar to nucleotides in the finished DNA
- DNA phosphate needs energy
Hydrolysis of extra phosphates provides energy for synthesis (from ATP)
makes the pair grow on the 3 prime side w ATP
the extra %%phosphates%% make up the phosphodiesther bonds on the sides
the released energy drives DNA synthesis
RNA Primase
%%RNA primase%% ==(4)==builds small complementary RNA primers at the replication fork on its own, these short pieces are called ^^RNA primers^^
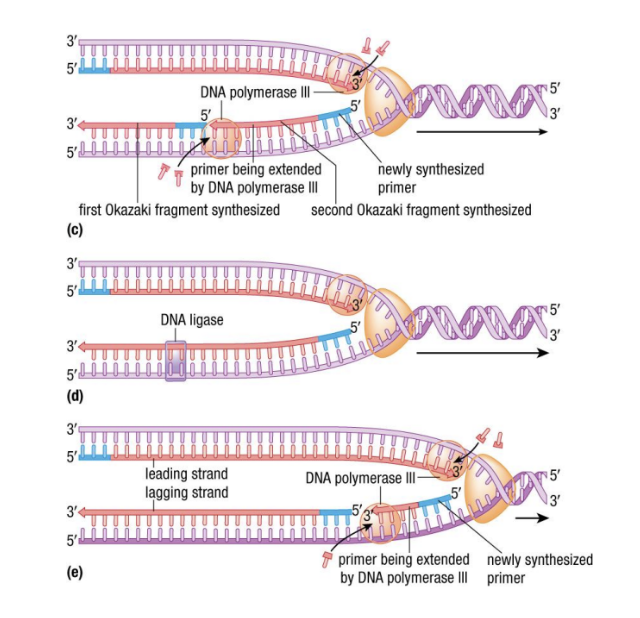
%%DNA polymerase III%% ==(5)== ( first one, last one discovered) synthesizes complementary strands in opposite directions (5’ → 3’), can only add to previously existing nucleotides
Leading & Lagging Strands
Leading strand is elongated toward the fork; efficient (5’)
^^Lagging strand^^ is elongated away from the fork, requires multiple RNA primers: constantly being synthesized backwards (3’)
- Results in segments of RNA primers & DNA called ^^Okazaki fragments^^
- %%DNA polymerase I%% ==(6)== replaces RNA primers with DNA - purifies
- %%DNA ligase%% ==(7)== joins Okazaki fragments into one continuous strand
<<Step 3: Repair<<
- Mismatched base pairs don’t bond properly and distort shape of the DNA molecule
- %%DNA polymerase II%% ==(8)== and other enzymes search for distortions, removes portion of the stand with mismatch and fills in the gap.