Gene Mutation, DNA Repair, Transposition
Vocabulary:
Mutation: An inherited change in the DNA sequence of genetic information, descendants that inherit this change may be cells or entire organisms, ultimate source of genetic variation and the source of may diseases/disorders
Germ-line mutation: A genetic mutation that occurs in a reproductive cell, only passed to half of offspring, but heritable
Somatic mutation: A genetic mutation that occurs in nonreproductive cells, passed to all new cells made via mitosis. Cannot be passed on, but can lead to altered cell function/tumors
Base substitution: A mutation where a single nucleotide base is replaced with another
Insertion: A mutation where material is mistakenly added
Deletion: A mutation where material is mistakenly removed
Point mutation: A base substitution from one base to another
Missense mutation: Point mutation that results in a new triplet code for a different amino acid
Nonsense mutation: A point mutation that results in a premature stop codon, terminates transcription early
Silent mutation: A point mutation where the new triplet code corresponds to the same amino acid, no functional effect
Transition: A base substitution where a pyrimidine/purine replaces itself (A/G and T/C)
Transversion: A base substitution where purine and pyrimidine are interchanged with one another
Frameshift mutation: A mutation that adds/removes a base pair, causes a shift in reading frame, altering all amino acids following the mutation site, can downstream produce a premature stop codon
In-frame insertion: A series of 3 insertion mutations, doesn’t impact the reading frame
In-frame deletion: A series of 3 deletion mutations, doesn’t impact ,the reading frame
Loss-of-function mutation: Mutation that reduces or eliminates functioning of a gene product
Null mutation: A complete loss-of-function mutation, results in complete loss of function
Dominant mutation: A mutation that when possessed on only one allele will result in a mutant phenotype, even if the wild type allele is still present
Dominant gain-of-function mutation: A mutation that results in a gene having enhanced, negative, or new function
Neutral mutation: A mutation that occurs in protein coding regions, where the effect on organism genetic fitness is neither beneficial nor detrimental
Spontaneous mutation: A change in nucleotide sequence that arises under normal conditions, results from DNA Polymerase errors, typically low rates for all organisms
Induced mutations: Mutations that result from influence of extraneous factors, like radiation, UV light, or natural/synthetic chemicals
Luria-Delbruck fluctuation test: Used in T1 phages, demonstrated mutations are spontaneous rather than adaptive
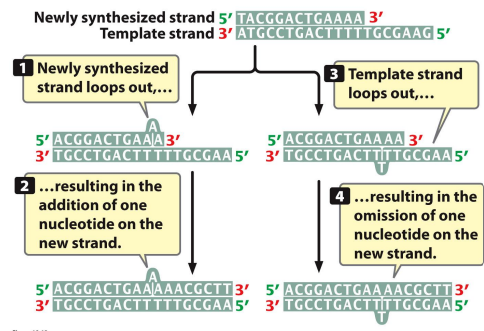
Replication slippage: When one strand of the DNA template loops out and is displaced, or when the DNA Polymerase slips/stutters during replication, more common in repeat sequences
Hot spot: Where lots of mutations may occur, due to many repeating sequences, contribute to hereditary diseases like fragile-X, Huntington’s, and myotonic dystrophy
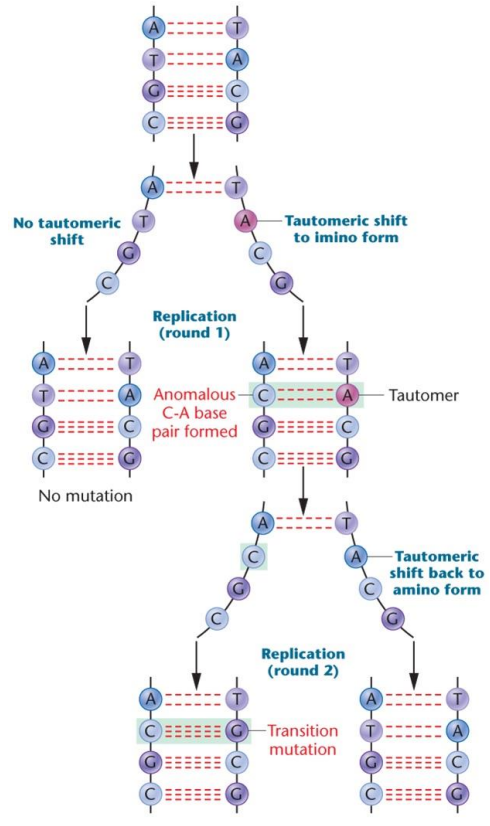
Tautomer: An alternate chemical form that differs by the shirt of a single proton in the molecule, has an increased chance of mispairing during DNA replication
Tautomeric shift: Can change the covalent bonding structure, allows noncomplementary base pairing and can lead to permanent base pair changes/mutations
Depurination: A mutation where a purine is lost
Apurinic site: A site on DNA where the helix is still intact, caused by depurination
Deamination: A mutation where an amino group is lost, happens to cytosine or adenine and causes A-T to turn to G-C
Mutagens: Natural or artificial agents that induce mutations above the spontaneous rate, includes things like fungal toxins, cosmic rays, UV light, pollutants, X-rays, and tobacco smoke chemicals
Oxidative damage to DNA: Damage that occurs because of normal cellular processes, exposure to high-energy radiator, super oxides, hydroxyl radicals, or hydrogen peroxide. Can produce over 100 chemical modifications to DNA including base modification, loss, and SSB
Transposable genetic element: Can move within or between genomes, present in all organisms, and can act as a naturally occurring mutagen to cause inversions, translocations, DSBs. Overall creates chromosomal damage
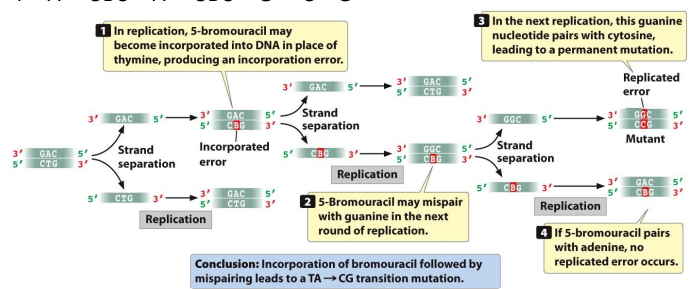
Base analogs: Can substitute for purines or pyrimidines during biosynthesis, tend to lead to increased tautomeric shifts and higher sensitivity to UV light
5-bromouracil (5BU): A base analog that behaves like thymine, alternatively has a Br in the 5 position. Can also pair with guanine when ionized, leading to a transition
Alkylating agents: Alter DNA, donate an alkyl group (CH3 or CH3CH3) to amino/keto groups, alter base pairing affinity and cause transition mutations
Intercalating agent: Different chemicals that due to their chape can wedge between DNA base pairs and cause distortions, unwinding, insertions, and deletions
Adduct-forming agent: Causes mutations by covalently binding to DNA to alter the conformation and interfere with replication and repair
UV Light: Part of the electromagnetic spectrum, at 260 nm is absorbed by DNA and damages it, tends to create pyrimidine dimers, used as a sterilizing agent
Pyrimidine Dimer: Caused by UV light, forms a bulky lesion that distorts DNA to block replication and inhibit cell division, causing cell death
Ionizing radiation: The energy of radiation varies intensely with wavelength, this penetrates deeply into tissues and ionizes particles, turning them into free radicals that break phosphodiester bonds, and overall causing chromosomal aberrations
Free radicals: A chemical species with one of more unpaired electron, can be created by radiation. These alter purines/pyrimidines, and can produce deletions, fragmentations, and translocations
Polygenic: Describes a disease caused by variations in several genes
Monogenic: Describes a disease caused by a single base-pair change
Beta-thalassemia: Autosomal recessive blood disorder, monogenic, less hemoglobin
Trinucleotide repeat sequence: A short DNA sequence found in a gene, normally may be low repeats but disorders can cause over 200 repeats
Strand discrimination: How different proteins know which strand is which, done base off of DNA methylation, new strand is unmethylated
Adenine methylase: A bacterial enzyme that recognizes DNA sequences and methylates adenine residues
Mismatch repair: Process that recognizes an unmethylated strand of DNA and can repair part of it, involves endonuclease, exonuclease, DNA Polymerase, and ligase
Endonuclease: An enzyme that nicks the backbone of newly synthesized DNA
Exonuclease: An enzyme that unwinds and degrades DNA after endonuclease acts on it, unwinding until a mismatch is reached
Post-replication repair: Repair to damaged DNA after it is replicated, done by RecA
RecA: A protein that directs recombination exchange with corresponding regions on undamaged strands
Photoreactivation repair (PRE): An enzyme that fixes pyrimidine dimers created by the introduction of UV light, activated by exposure to visible light by absorbing a photon, uses this energy to cleave the thymine dimer, found only in e. coli
Excision repair: When an exonuclease recognizes and cuts errors, DNA polymerase will insert complementary nucleotides in the gap and ligase will seal the bonds
Base excision repair (BER): Excision repair that corrects DNA with damages bases, uses DNA glycosylase to recognize the altered base
Nucleotide excision repair (NER): Excision repair that repairs bulky lesions that alter/distort the double helix, excises 13 nts in prokaryotes and 28 nts in eukaryotes
DNA Glycosylase: 4 different types, one for each base, used in BER, recognizes an altered base pair
uvr gene product: Will recognize and clip out lesions in NER
Xeroderma Pigmentosum (XP): A disorder where proteins in the NER system are nonfunctioning, have 2000x chance of developing cancer, individuals have skin abnormalities and are sensitive to light/UV radiation
Cockayne syndrome (CS): Disease of the NER system, has developmental and neurological defects, premature aging and death ~20, sunlight sensitivity
Trichothiodystrophy (TTD): Autosomal recessive disease of the NER system, causes dwarfism and intellectual disabilities, sensitivity to sunlight and a 6 year life span
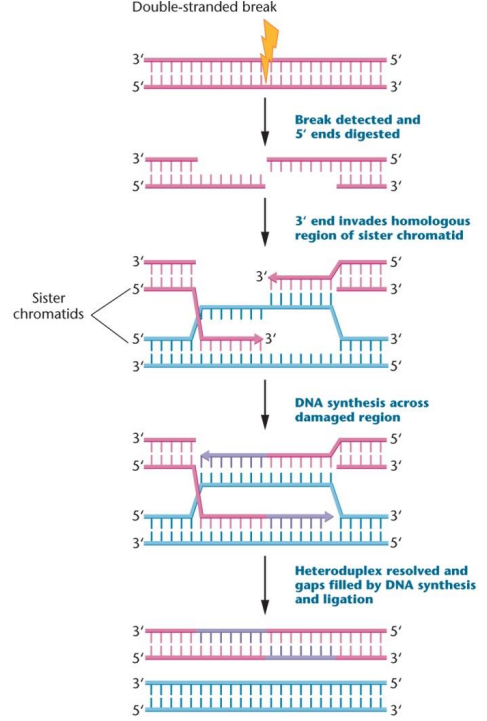
Homologous recombination repair pathway: Recognizes a break and digests the 5’ end (leaving 3’ overhang), 3’ end aligns with complementary sequence on sister chromatid, and DNA synthesis starts from the undamaged DNA strand as a template, occurs late S or early G2 phases
Nonhomologous end joining pathway: Occurs during G1 to repair DSBs, includes a complex of proteins including kinases and BRCA1, they may bind to free ends of DNA to trim and ligate them back together, error prone as some bases are lost, if more than 1 chromosome breaks potential for Robertsonian translocation is possible
Ames test: Uses strains of salmonella typhimurium to reveal presence of certain mutations, an assay measures a chemical’s toxicity by its ability to induce reverse mutations in a mutant gene, used to develop pharmaceutical chemical compounds
Transposon: A transposable element, “jumping gene” that can move between chromosomes, found in all organisms with unknown precise function
Autonomous transposon: A transposon that can transpose itself, encodes functional transposase enzymes
Nonautonomous transposon: A transposon that cannot move itself, doesn’t encode its own transposase and require presence of autonomous transposons elsewhere in the genome
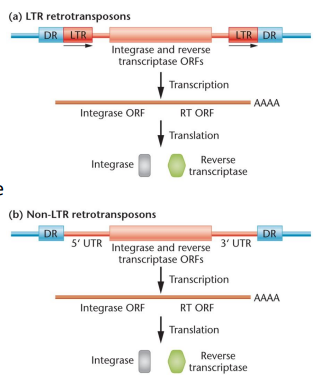
Retrotransposon: A transposable element that amplifies and moves itself within a genome, uses an RNA intermediate, can resemble a retrovirus, can be autonomous/nonautonomous, long-terminal-repeat/nonLTR, have an ORF and encode reverse transcriptase
Human transposable elements: Half of the human genome, consists of long interspersed elements (LINES) and short interspersed elements (SINES), which are non-LTR retrotransposons
With the Luria Delbrück fluctuation test, there were 2 hypotheses. If mutations were induced, there would be a constant rate of mutation and when different strains of T1 phage were exposed, each plate would contain roughly the same number or resistant colonies. If it was random, mutations could occur in the absence of T1 with a low fixed rate of resistance is mutations occurred late. If mutations occurred early, overall fluctuating numbers in spontaneous populations from generation to generation would stay constant. Data supported the second theory
E. Coli will repair DNA only when minimizing DNA damage and in the presence of large numbers of mismatches/gaps, synthesis becomes prone to errors, can be mutagenic allowing cells to mutate and survive with damage
With retrotransposons, a cell will use its natural RNA polymerase to transcribe them, which when translated will go on to make more copies of the gene through reverse transcription to integrate their DNA back into the cell, causing more transcription in positive feedback. They tend to create mutations at many sites in the genome, and act as copy/paste due to the original retrotransposon not being excised