M2L1 Epigenetics and genomic landscapes
Functional elements in linear sequences
Core promoter
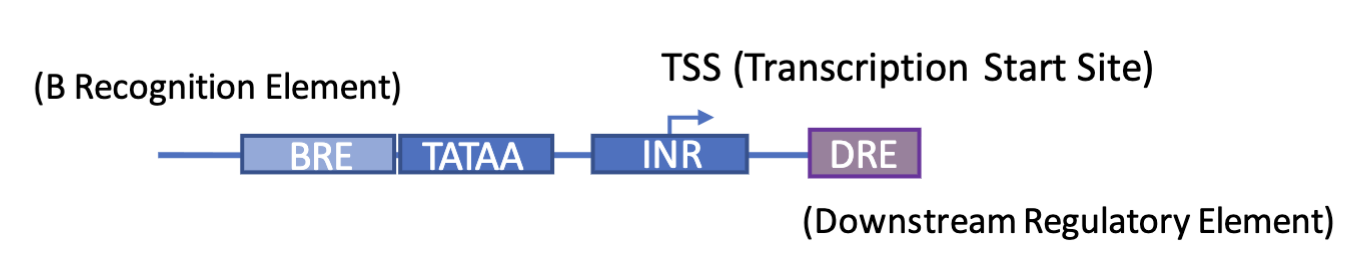
Transcription start site (TSS) - where transcription is initiated
TATA box - AT-rich core promoter element (binding site for TFs)
TFIID complex contains TATA-binding protein (TBP) to bind to TATA and recruit general transcription factors and RNA pol II
B recognition element (BRE) - flanks the TATA box and is reognised by TFIIB which bridges TBP and RNA pol II
Downstream regulatory element (DRE) - recognised by components of TFIID, particularly TBP-associated factors (TAFs) which enhance transcription
Upsteam regulation
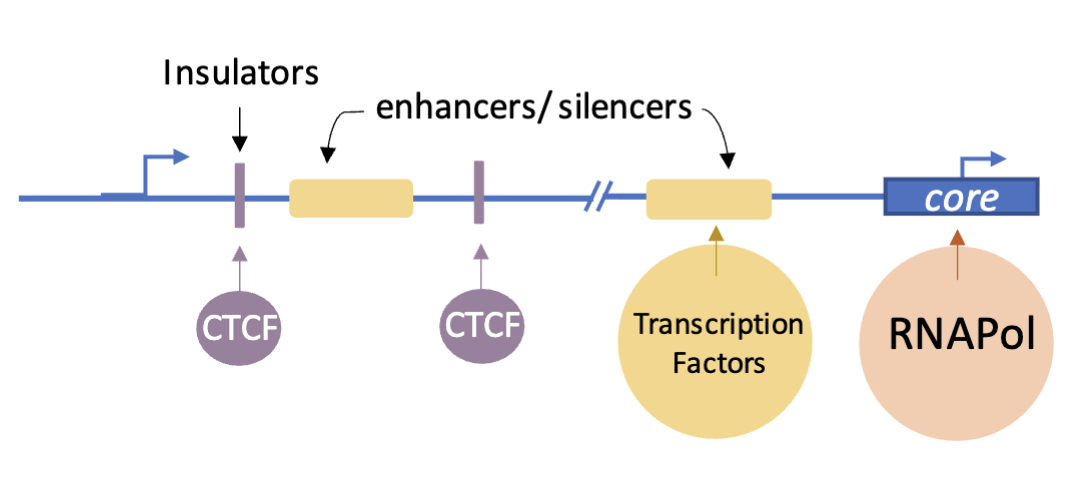
RNA pol binds to the core
Transcription factors may bind upstream, very close or also distally to the core (relying on DNA looping to contact the RNA pol)
More transcription factors interacting with RNA pol results in greater transcription
Insulators - boundary. elements that block interactions between enhancers and promoters when positioned between them
Bound by CTCF (CCTC-binding factor), forming chromatin domains and ensuring that an enhancer only activates the correct target gene and not neighbouring ones
Distance requlation
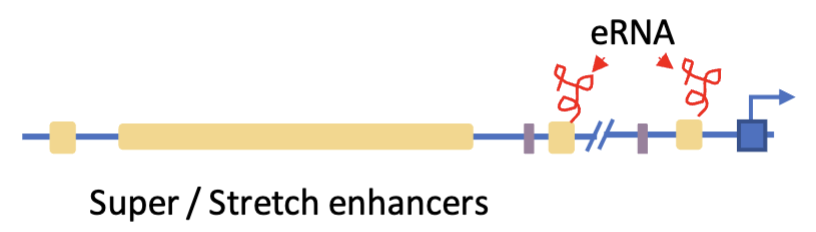
Super/stretch enhancers - massively upstream but can be looped to make contacts with distant regions
Enhancer RNAs (eRNA) - involved in gene regulation
Splicing
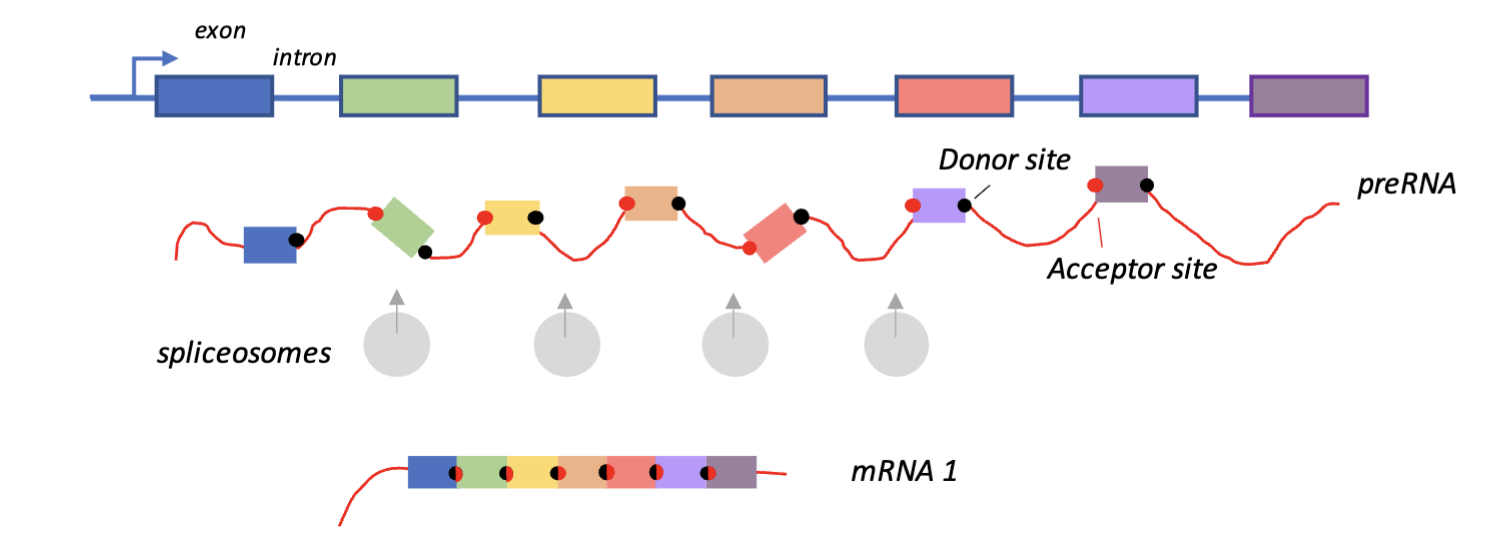
Introns and exons can be spliced to produce mature mRNA (removing introns)
Splice sites and acceptor sites are recognised by spliceosomes
Splice sites can be mutated in cancer, disrupting splicing patterns leading to inclusion of introns - they may have stop sequences and cause abortive translation
Exon skipping
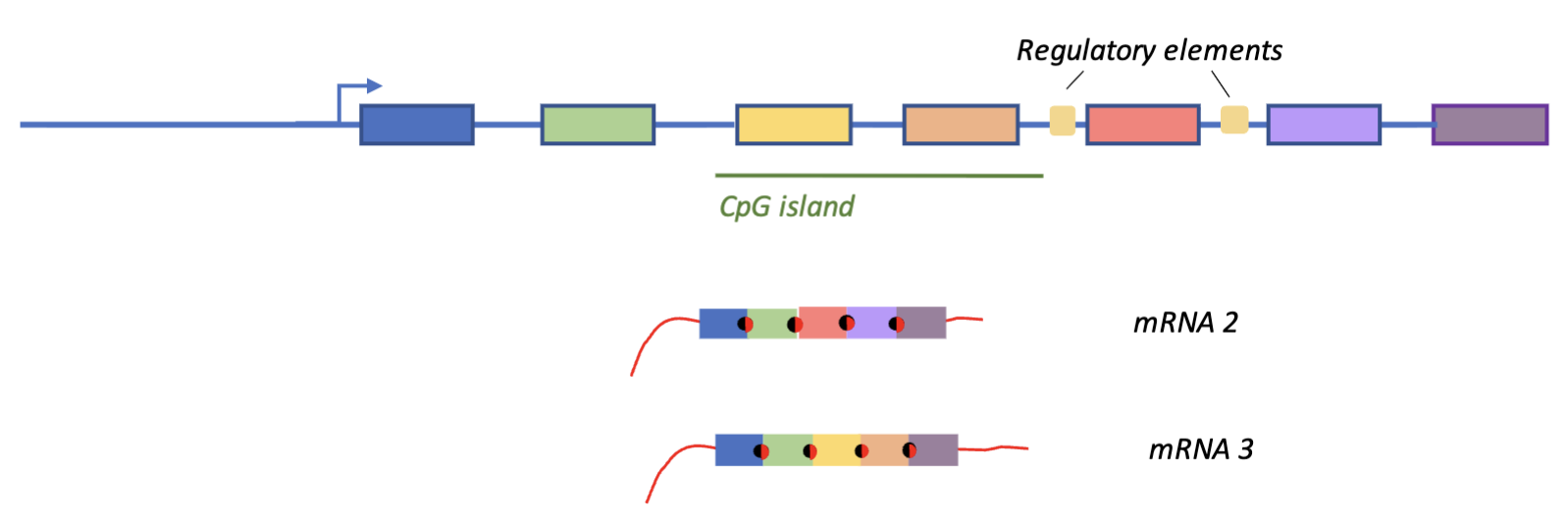
DNA methylation in CpG island might silence some exons, causing them to be skipped when splicing exons together
Regulatory elements may cause single exon skipping
Alternative promoters
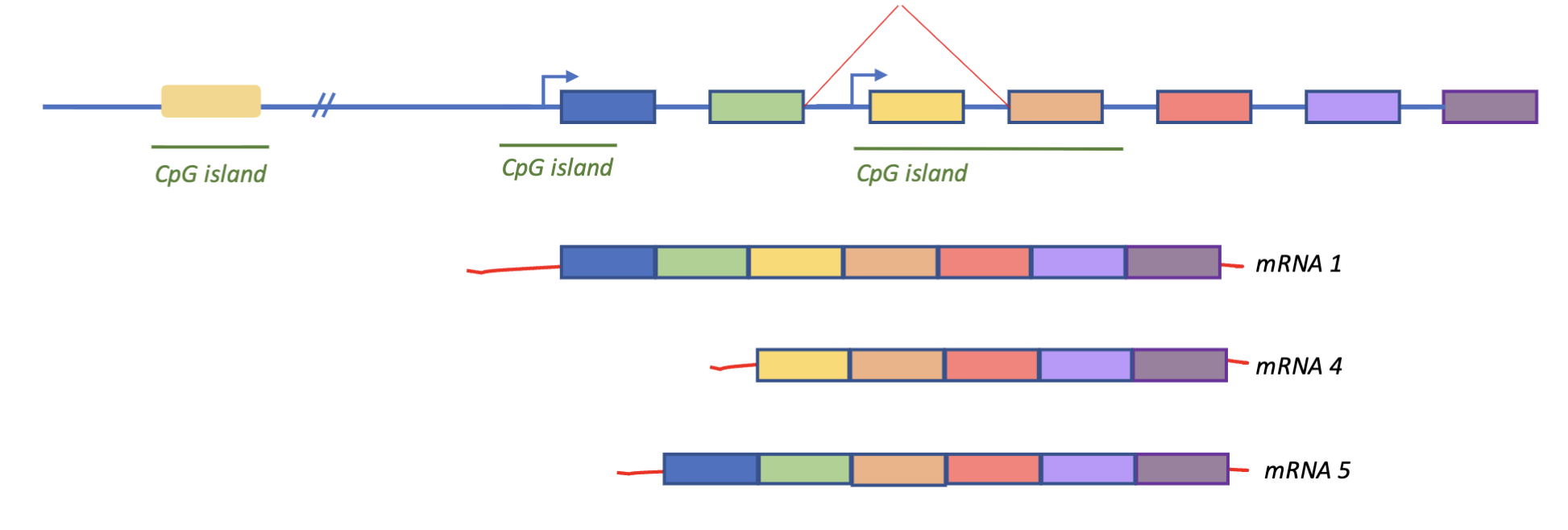
CpG islands can sit around core promoters, enhancer elements, or internal in the gene (internal core promoter)
RASSF1 bivalent promoter of solid cancers (Hippo tumour suppressor pathway)
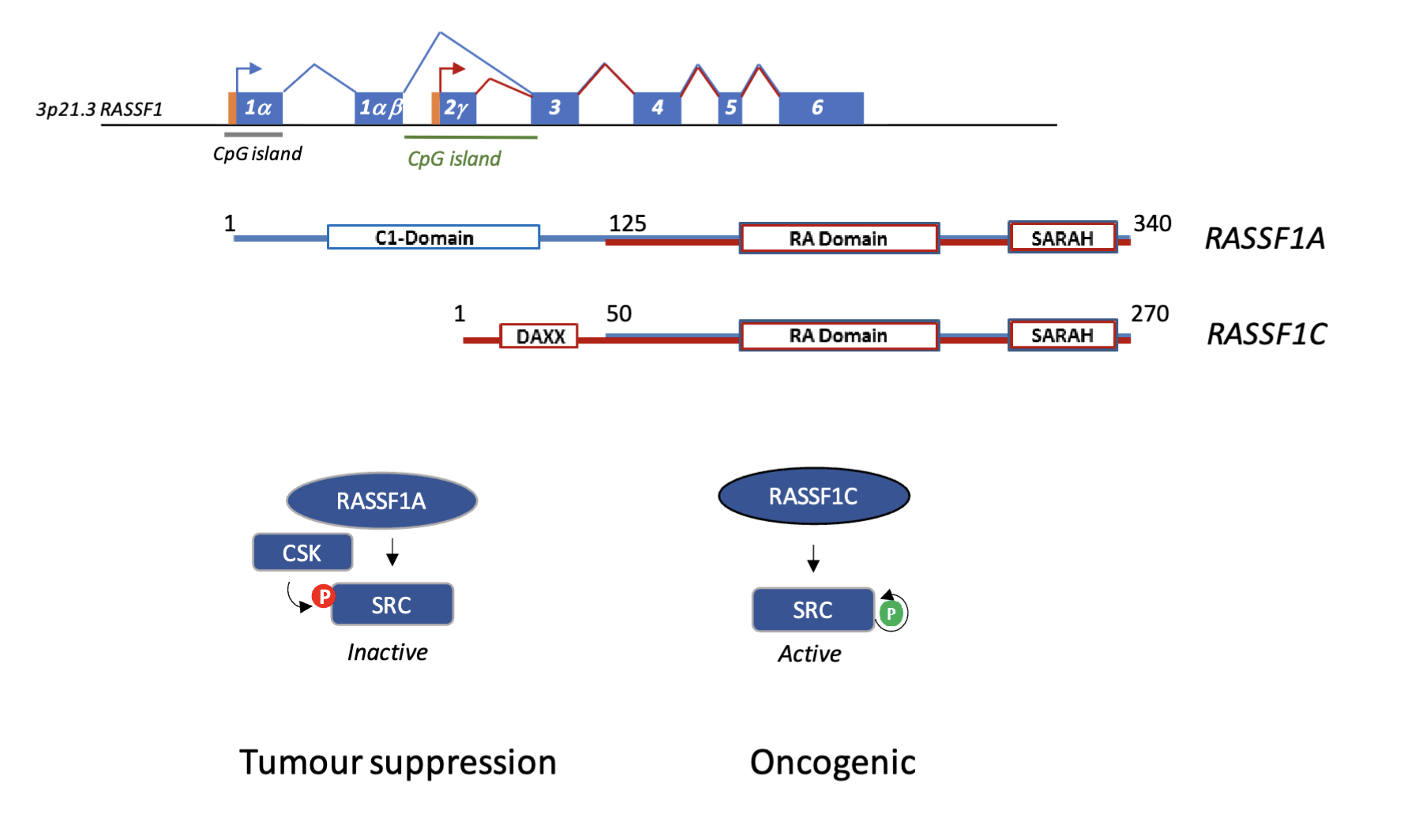
1α CpG island is in the promoter region of RASSF1A and and the 2γ CpG island is in the promoter of RASSF1C
When 1α is methylated transcription starts from 2γ, producing the C isoform and when 2γ is methylated transcription starts from the 1α promoter, skipping 2γ to produce the A isoform
Two different transcripts can be produced - RASSF1A or RASSF1C
Both have identifical C terminal, Ras associated (RA) domain, and SARAH domain (protein-protein interaction domain)
A isoform has a unique N terminal
A and C can both bind to/activate Src using their SARAH domain
A isoform can also bind to C terminal Src kinase (CSK) which inhibits Src, whereas C isoform does not bind to CSK and only activates Src (Hippo signalling)
RASSF1 alternative promoters impact prognosis
High methylation in RASSF1A / low methylation in RASSF1C = worse prognosis
Translational control
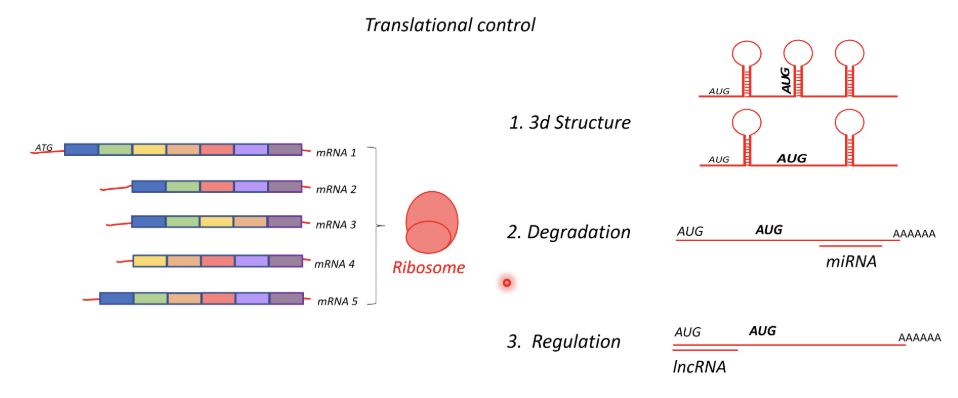
AUG is the start sequence - presence of stem loop structures within transcripts can influence where the ribosome starts translating if there is AUG in the middle of the transcript
Eg. AUG at the beginning may initiate transcription if AUG in the middle is ‘hidden’ in a stem loop strutcure, but if this structure is disrupted translation may start in the middle using this as an internal ribosomal start side
Degradation by miRNA or siRNA - target RNA degradation
Can regulate transcription depending on what is needed at the time, eg. if a short term rapid response is needed for an acute environmental trigger, a burst of transcription may be needed for a short time, after which it may no longer be needed, alternatively for long-term processes like development transcripts may need to be produced slowly and stably over longer periods
Regulation by lncRNA - stop the reading of transcripts
Chromosome organisation
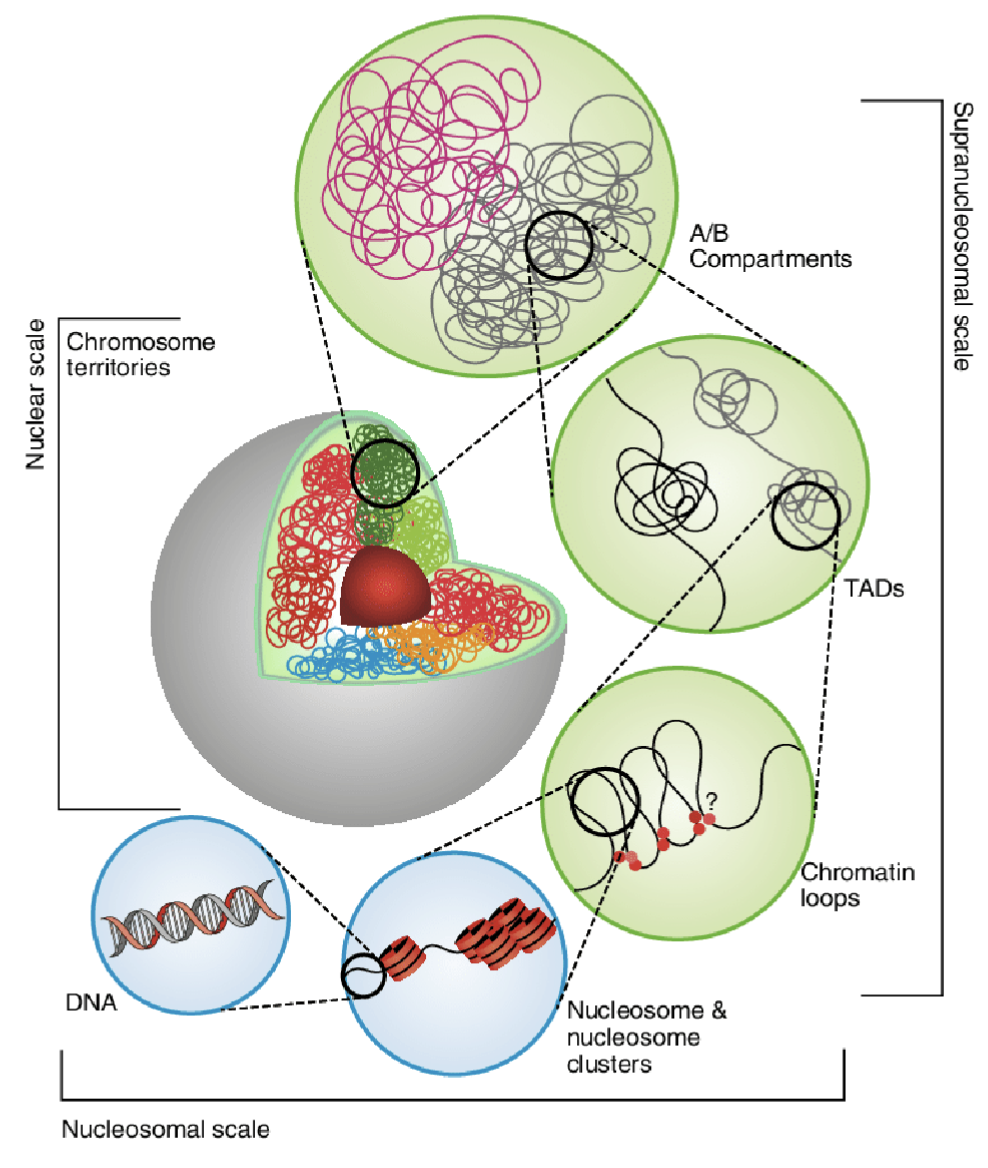
Each chromosome occupies (territory) its own space in the nucleus
At the periphery of the nucleus there is more repressive machinery which silence genes (B compartment), whereas in the centre there is more open chromatin and active genes (A compartment)
Topologically associated domains (TADs) - interacting aspects of different chromosomes in 3D space
Chromatin loops - DNA strand loops to bring together distant genomic regions (eg. enhancer and promoter), typically smaller and more specific interactions occuring within (or sometimes between) TADs
In between territories there are:
Nuclear speckles, PML bodies, cajal bodies, and paraspeckles - different types of nuclear bodies
Nuclear speckles - storing and supplying RNA splicing factors
Cajal bodies - assembling and modifying snRNPs and snoRNPs for RNA splicing and rRNA modification
PML body - contains PML protein and regulatory proteins for DNA repair, apoptosis, antiviral response, senescence and tumour suppression
Paraspeckles - retaining RNA and RNA-binding proteins in stress adaptation, differentiation, and stress regulation
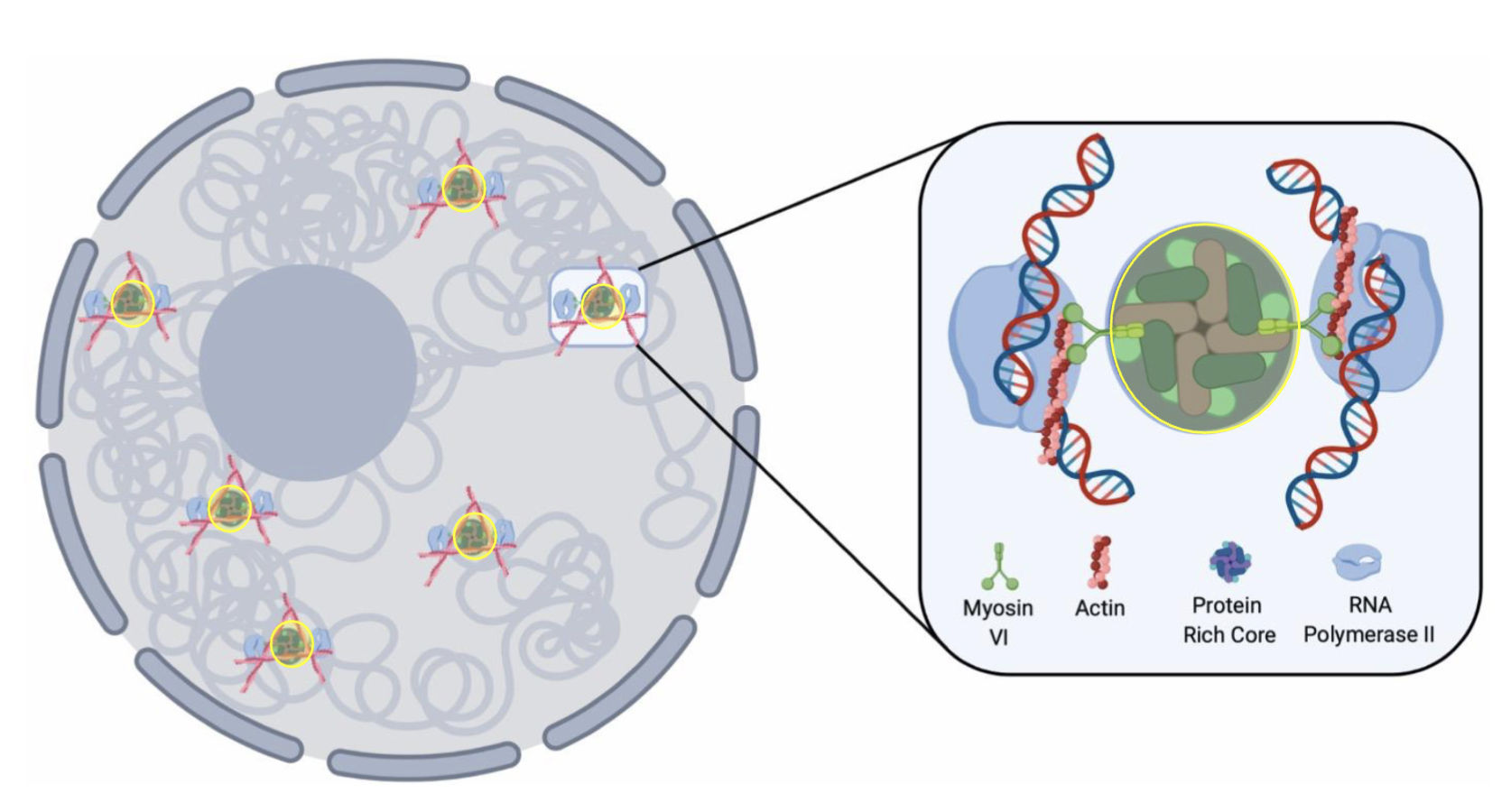
Splicing factories
Transcription factories
Polycomb domain
Transcription factories are physically confined by nuclear actin to give stability, allowing them to go through the different stages of transcription
Nuclear actin and myosin are associated with phase separation factors - driven by hydrophobicity/hydrophilicity of proteins
Interface of phase separated boundaries are a site of transcription
Transcription factories
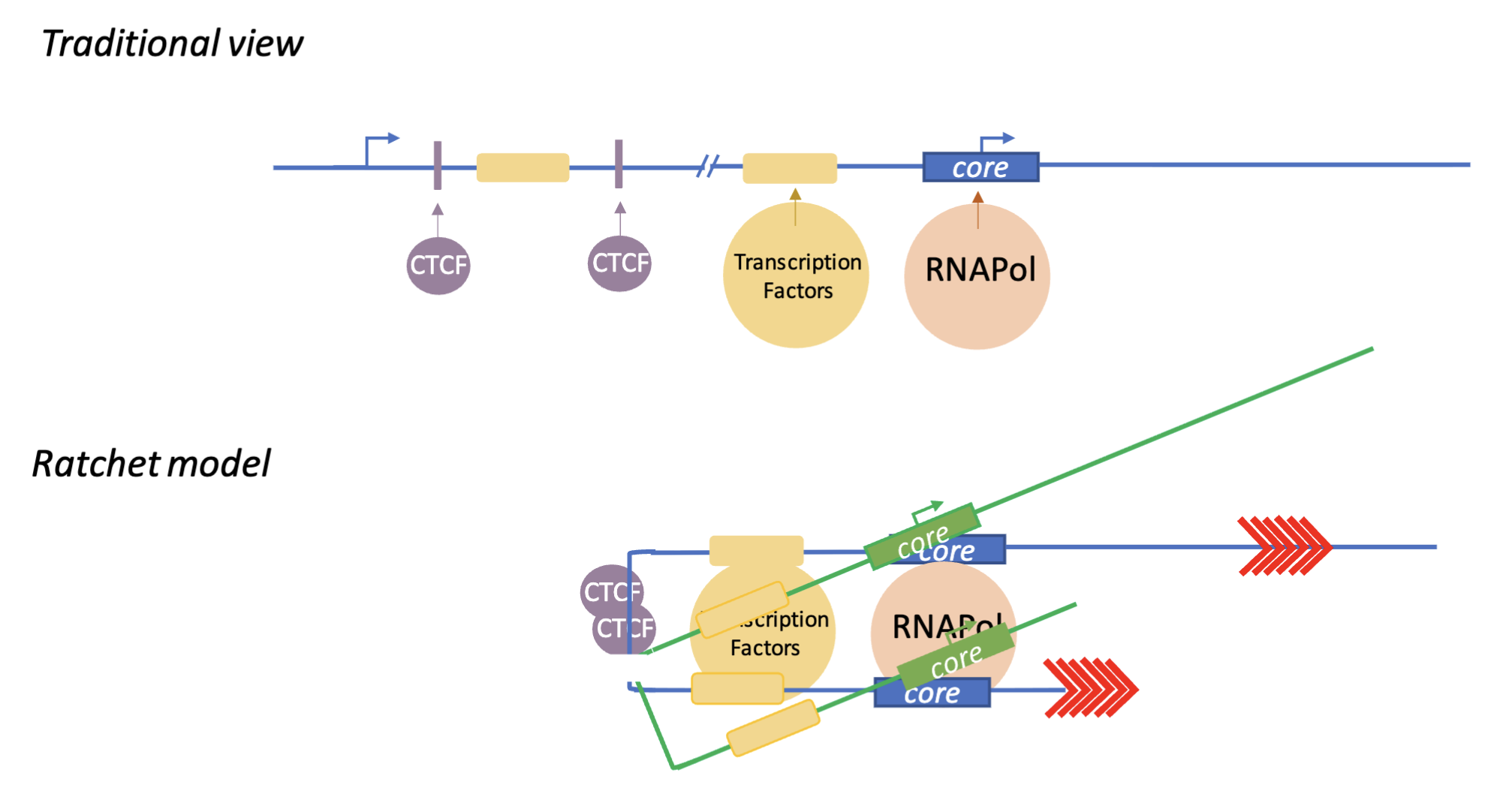
Ratchet model
Circularisation of genes - transcription factors just need to be activated in one place and necessary genes can be ‘dragged in’ via chromatin looping
Multiple genes can be activated at once
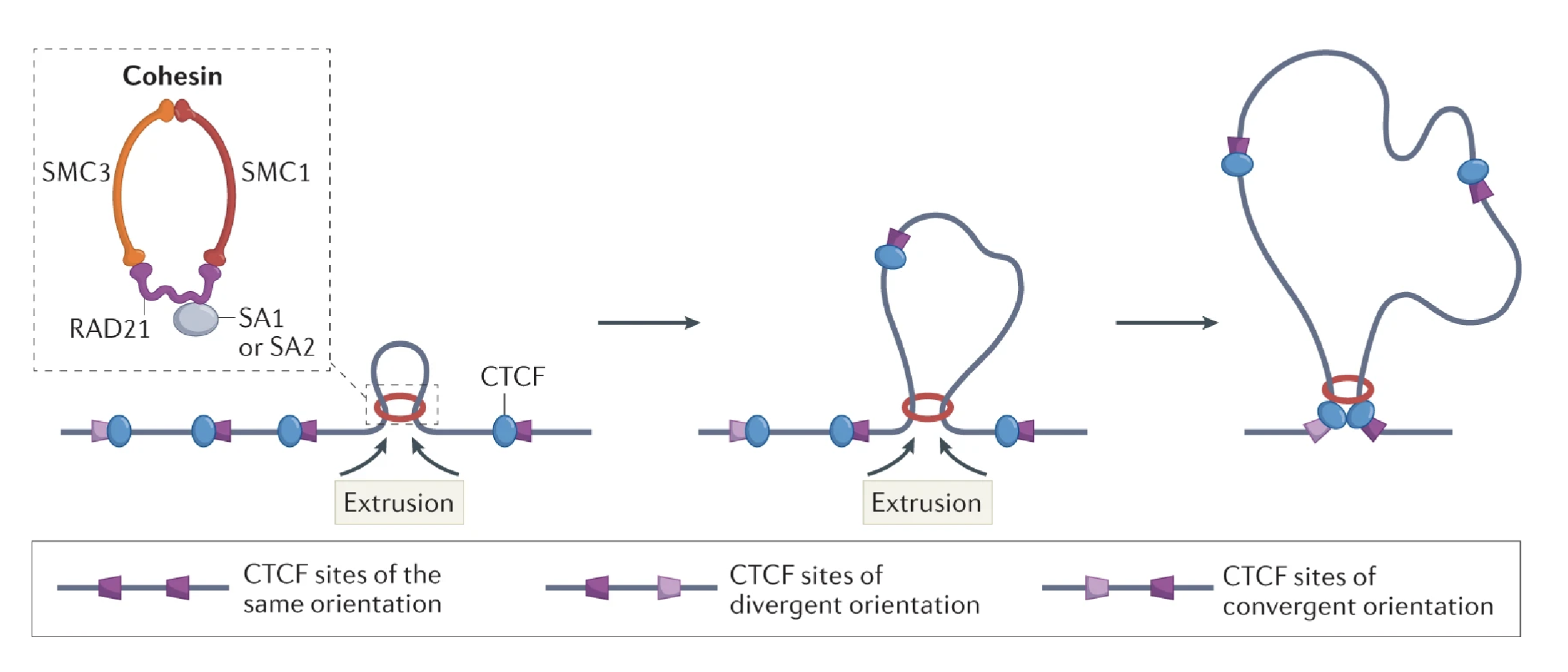
DNA is extruded through cohesin rings, bringing CTCF sites together, oriented either in the same or opposite direction
CTCF sites in the convergent orientation stops the DNA from looping out
Mediator complex - scaffolding of TF complex to facilitate their stability
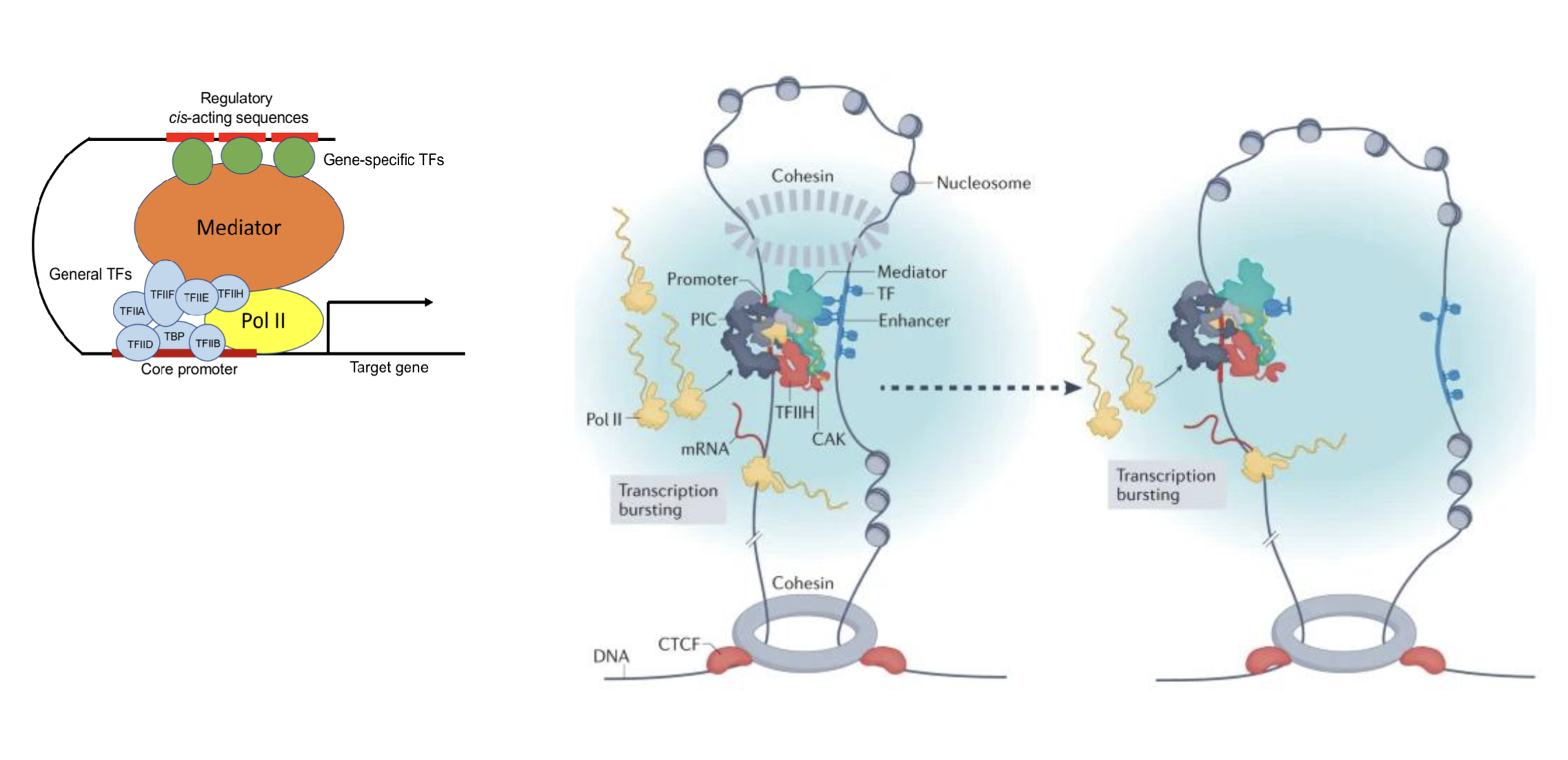
Mutations do not directly cause cancer, eg. we all have mutations due to environmental factors like UV etc but not all of us have cancer, it is likely due to mutations in mediators and regulators of gene expression that cause the mutations to become visible
Concentrates transcription machinery in a region, allowing for transcription re-initiation and rapid cycling of the process
Topological associated domains (TADs)
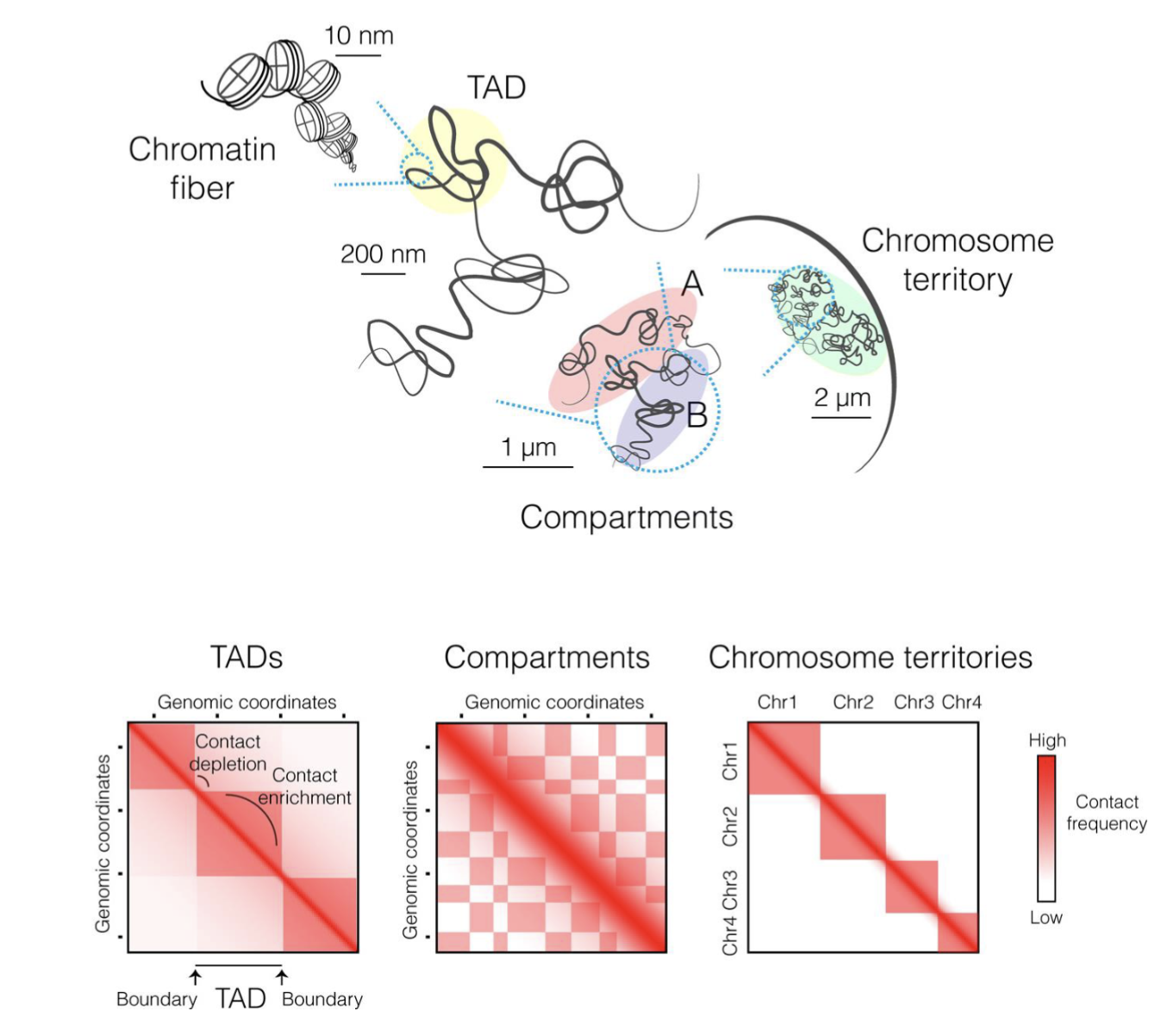
Using chromosome conformation capture methods (CCC) - interacting regions of chromatin can be crosslinked using formaldehyde, DNA is then digested and remaining fragments are ligated and DNA is analysed after removing crosslinks
Sequencing of interacting regions reveals the presence of TADs, compartments and chromosome territories
TADs are large chromosomal regions in which DNA sequences interact more frequently with each other than sequences outside the domain
Act as functional neighbourhoods - enhancers within a TAD tend to regulate promoters within the same TAD
Insulates gene regulatory activity, preventing enhancers from activating genes in neighbouring TADs
Lamin associated domains (LADs)
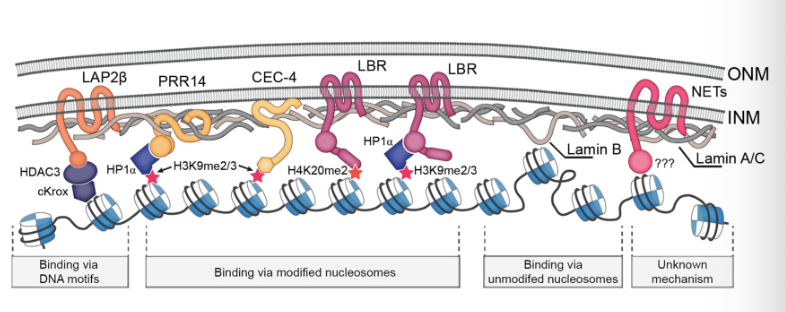
Lamin is the structural component inside the nuclear envelope, forming a rigid structure which protects the genome
Chromatin that is closest to the nuclear envelope are LADs
Proximity of histones and chromatin modifiers (histone deacetylases, methylases) are lined up in association/bound with proteins in the inner leaflet of the nuclear envelope
Presence of repressive machinery at the inner leaflet causes genes within LADs to be silenced
LADs position chromatin at the nuclear periphery, organising the genome into the active central compartment and inactive peripheral zones
During development or differentiation LADs can detach or form, allowing changes in which genes are active or silenced
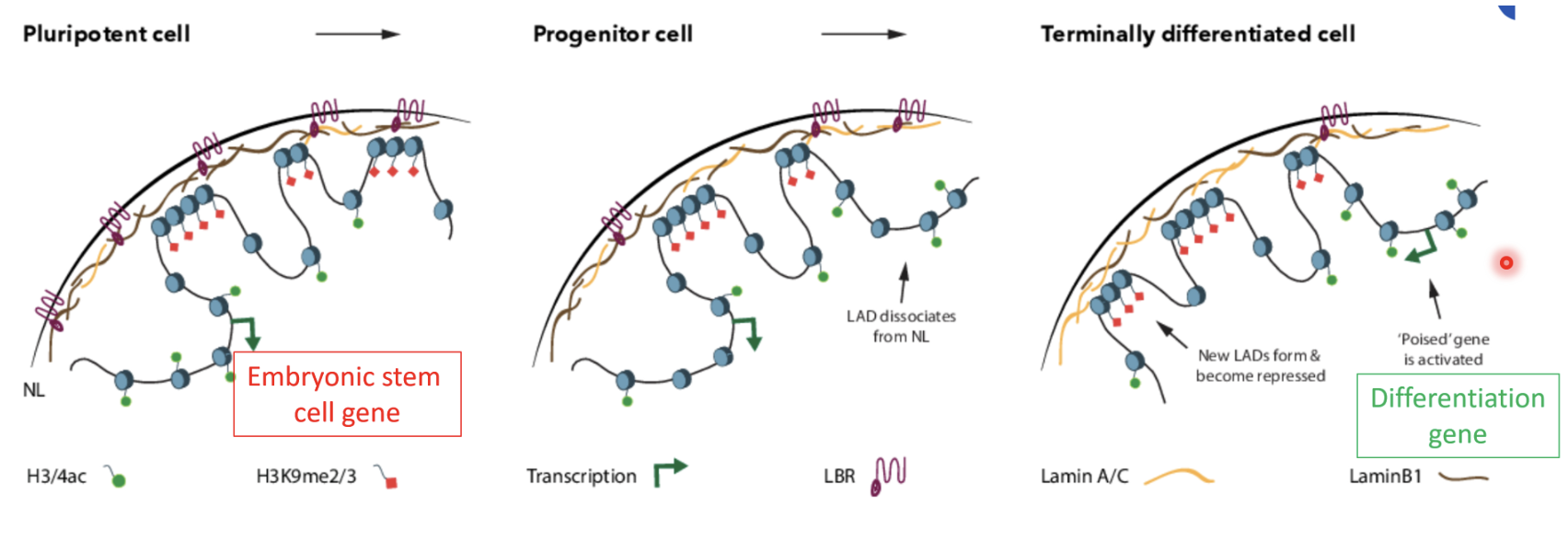
Nucleosome remodeling and deacetylase complex (NuRD) and polycomb repressive complex 2 (PRC2) are the epigenetic core of LAD silencing
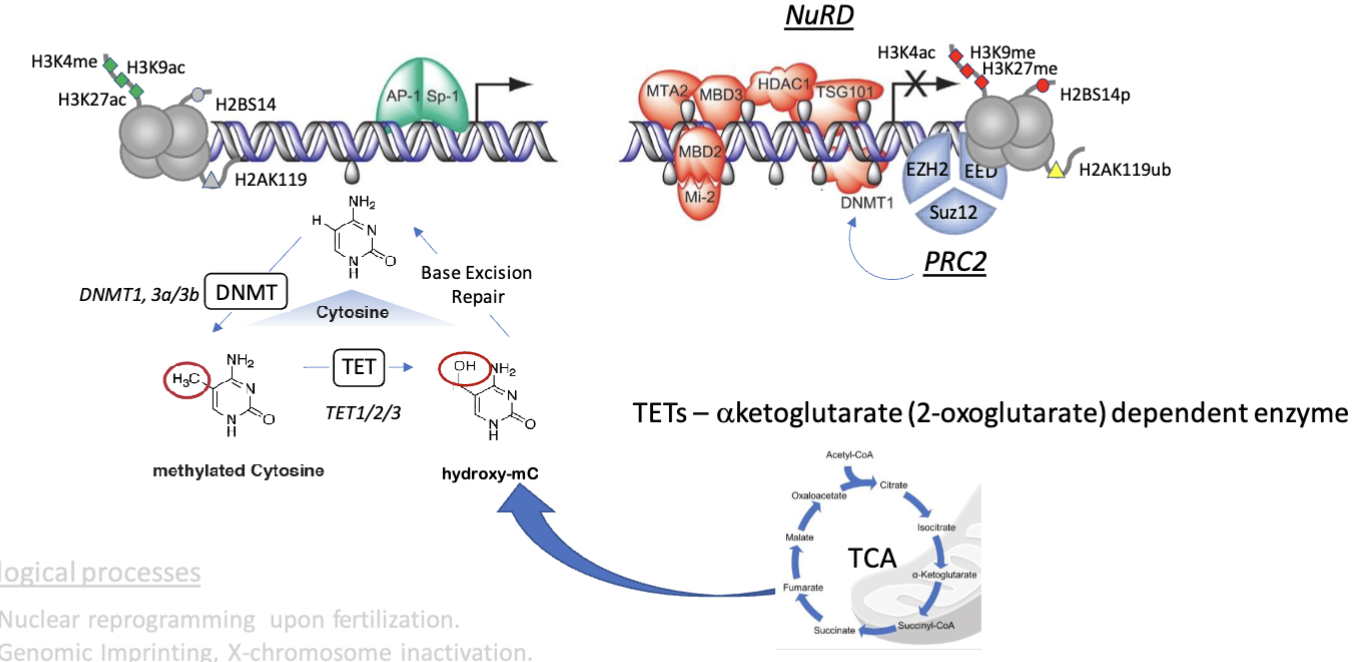
This is implicated in nuclear reprogramming upon fertilisation, genomic imprinting, X chromosome inactiation, development, wound repair, dedifferentiation in cancer
5hmC tends to be at the periphery of CpG islands whereas 5mC was in the middle
5hmC is associated with transcriptionally active or poised promoters, functioning as a transmition mark to either maintain a dynamic, active chromatin state or marking sites being demethylated by TET enzymes
5mC is common in repressed promoters, such as those bound by Polycomb complexes or in LADs, which recruits MBD proteins which further recruit NuRD and HDACs to enforce repression
5hmC tends to decrease in cancer progression causing global hypermethylation and transcriptional silencing
TETs are dependent on α-ketoglutarate (a TCA cycle intermediate), hence decreasing 5hmC reflects deregulated cellular metabolism/shift to aerobic glycolysis
Nucleolar associated regions (NADs)
The nucleolus contains rDNA for ribosome synthesis
Chromosomes with rDNA components ‘stick their head’ into the nucleolus
There are hundreds of copies of ribosomal genes to make sure we can make enough
>95% of cells energy goes into making ribosomes
Nucleolus also has lamin A/C - continuous link between the envelope and the nucleolus as this would allow ribosomes to get out into the cytoplasm easier
rDNA units are lost over time as they are heavily transcribed
Instead of repairing damage in this region it is removed, since there are hundreds of copies of these genes
Safer to lose the gene then trying to repair it because they are highly repetitive
Associated with biological aging
Different from chronological age associated with telomeres as telomeres are non-functional, whereas rDNA is functional and alcohol consumption, smoking, stress etc will require more repair proteins to be produced which uses up more units
Causes of cancer
Genetic - eg. mutations in p53 (Li-Fraumeni), BRCA1/2 (breast and ovarian cancer), NF1/2 (neurofibromatosis)
Environmental - eg. smoking and lung cancer, asbestos in mesothelioma
Sporadic - ‘dark genome’?
Mutations in genes - inactivating (nonsense deletion, stop, frame shift) in tumour suppressors, activating (change protein function) in oncogenes
Mutations that affect gene regulation - gene activity, translation, genomic topology
Chromosome translocation - more likely between chromosomes in neighbouring terrotories or sharing the same transcription factory
Form follows function
Mutation/translocation, altered methylation, altered CTCF
—> Loss of neighbourhoods —> Altered gene expression