Enzymes
Mechanisms
Enzymes are biological catalysts. They speed up reaction rates by reducing the activation energy of a given reaction. Enzymes are also sensitive to temperature, pH, and are specific for certain reactions or classes of reactions.
Enzymes are not used in the reaction and do not affect thermodynamic parameters, or the equilibrium of a reaction. Because they do not affect thermodynamics- they cannot turn an energetically unfavorable reaction into a favorable one (unless they couple reactions together).
Enzymes do:
increase the rate of reaction
reduce the activation energy
sensitive to pH and temperature
are specific to certain reactions or reaction classes
Substrate: the substance that the enzyme operates on
Active site: the space in which enzymes and substrates interact (can be separated into a binding site and a catalytic site)
Four catalytic strategies that enzymes can use are: acid/base, covalent, electrostatic, and proximity/orientation.
Two models can describe how enzymes interact with substrates:
lock and key: the active site of an enzyme and the substrate fit together like puzzle pieces with no change in tertiary or quaternary structure
induced fit (the now most accepted): substrate and enzyme affect each other by inducing changes in their respective tertiary and quaternary structure. Allows for closer binding and more efficient catalysis
Both models have enzymes that are substrate specific.
Classification
The main enzyme classes are as follows:
Hydrolase
hydrolyzes chemical bonds (includes ATPases, protease, lactase)
Isomerase
rearranges bonds within a molecule to form an isomer (ribose-5-phosphate isomerase)
Ligase
forms a chemical bond (DNA ligase, pyruvate carboxylase)
Kinase
transfers a phosphate group to a molecule from a high energy carrier, such as ATP (e.g PFK)
Oxidoreductase
runs redox reactions (includes oxidases, reductases, dehydrogenases, and others)
Polymerase
polymerization (addition of nucleotides to the leading strand of DNA by DNA pol III)
Phosphatase
removes a phosphate group from a molecule
phosFATtase- it needs to lose WEIGHT so it removes a phosphate group
Phosphorylase
transfer a phosphate group to a molecule from inorganic phosphate (laces a phosphate to whatever group its being added to)
Protease
hydrolyzes peptide bonds (trypsin, chymotrypsin, pepsin, etc.)
Regulation
There are many ways in which enzyme activity can be regulated which includes:
Covalent modifications
Proteolytic cleavages (also a covalent modification)
Associations with other polypeptides
Allosteric and Osterosteric regulation
Feedback regulation
Covalent Modification
This involves the addition of groups via covalent bonds that can regulate enzyme activity, enzyme lifespans, and/or cellular location.
Acetylation: addition of an acetyl group
Methylation: addition of a methyl group
Glycosylation: addition of a glucose molecule
Suicide inhibition: is an inhibitor that covalently bonds to an enzyme to permanently prevent it from catalyzing reactions
Proteolytic cleavages
Some enzymes could be within their inactive forms called zymogens (or proenzymes), which requires proteolytic cleavages in order to activate the enzyme.
Sometimes enzymes requires a helper molecule to assist biological functionality. These help molecules are called cofactors and can be either inorganic or organic.
organic factors can also be called coenzymes
coenzymes that are tightly or covalently bonded to their enzyme are called prosthetic groups
holoenzyme= enzyme+ cofactors
apoenzyme (apoprotein)=enzyme without cofactors
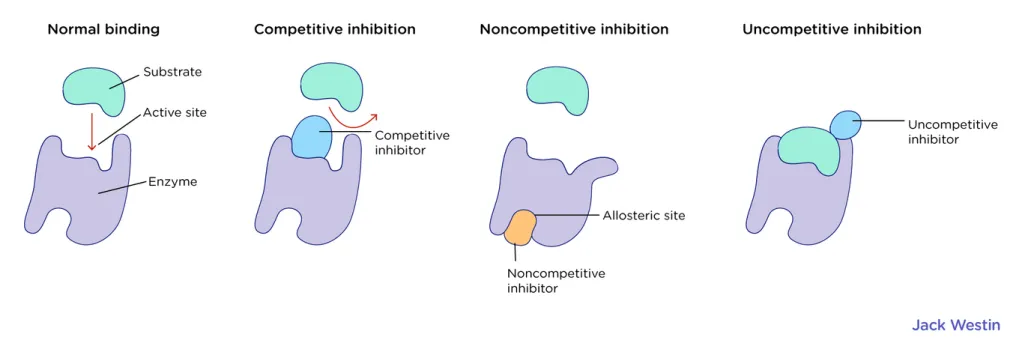
Allosteric and Osterosteric regulation
Regulators can either inhibit or activate enzymes and can either be bound osterosterically (within the active site) or allosterically (in a site other than active site)
heterotrophic regulator: a regulator that is not also the substrate
homotropic regulator: a molecule that is bother a substrate and regulator
Most common regulations of these types are competitive, noncompetitive, and uncompetitive inhibitors.
competitive inhibitor: an inhibitor that binds on the active site of an enzyme
noncompetitive inhibitor: an inhibitor that can bind to the allosteric site (or binds to the enzyme-substrate complex) either before or after the substrate is bound. Does not prevent substrate from binding to active site
uncompetitive inhibitor: bind to an allosteric site (away or near the substrate site) and prevents the substrate from leaving
mixed inhibitor: is a mix of multiple types of inhibitors
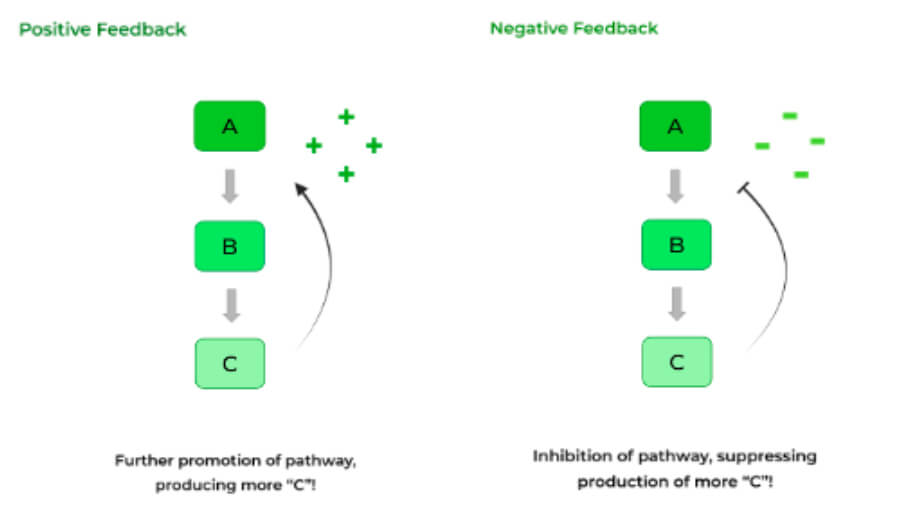
Feedback regulation
Enzymes are commonly regulated by downstream products which is called feedback regulation.
There are two main types:
negative feedback/feedback inhibition: downstream products or molecules downstream goes to inhibit the enzyme further up the loop.
positive feedback/feedback stimulation: products downstream causes the enzyme upstream to be further stimulated (oxytocin and uterine contractions).
Kinetics and Inhibition
Enzymes can be regulated through inhibition. Certain inhibitors affect difference aspects of enzyme kinetics based on how it binds and how it alters the enzyme.
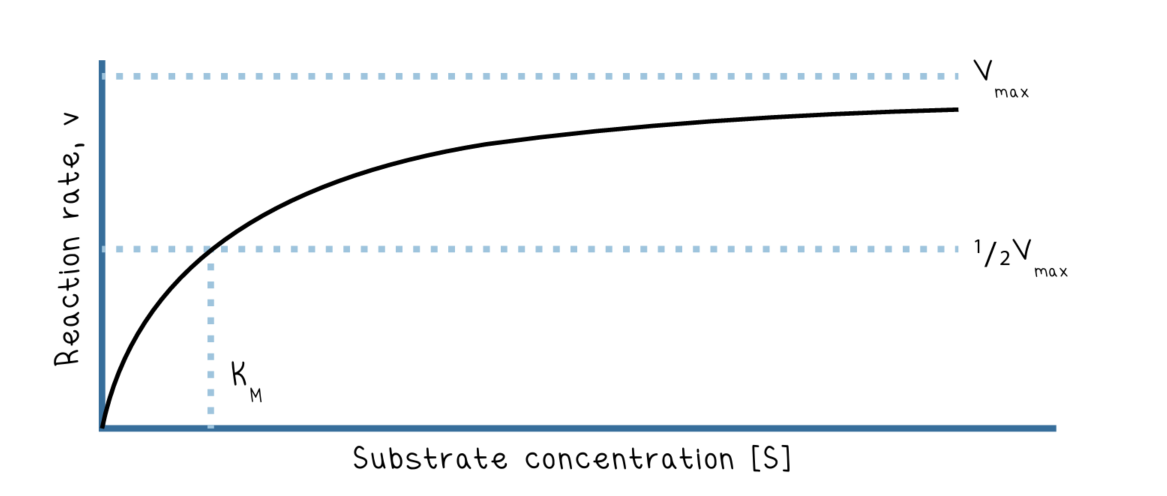
Michaelis-Mention model of enzyme kinetics:
a model that shows the affects of enzyme inhibition through a reaction rate (V) vs substrate concentration graph (S)
The slope of the model is nonlinear because eventually the enzymes present will become saturated or occupied.
the equation for Michaelis-Mention is: V0= (Vmax*[S])/(Km+[S])
Variables:
V0: the initial velocity of the reaction (unit is s^-1)
depends on the concentration of substrate and enzyme, and is directly proportional to the amount of substrate (if enzymes aren’t fully saturated)
Vmax: maximum rate of the reaction (unit is s^-1)
when enzymes are fully saturated with substrate
[S]: concentration of substrate (in micromolar)
Km: the Michaelis constant, the concentration of substrate which permits the enzyme to achieve half of Vmax
when the substrate concentration when the reaction velocity is half its Vmax (km=1/2Vmax)
it is also the inverse measure of affinity (higher km=lower affinity). Enzyme works better when there is a lower Km (meaning the enzyme has a higher affinity for the substrate)
Kcat: enzyme turnover number
how many substrate and enzyme can turn into a product per one second at its maximum speed (units are sec^-1)
solved by solving kcat=Vmax/Et
Et= total concentration of available enzyme
catalytic efficency= kcat/km
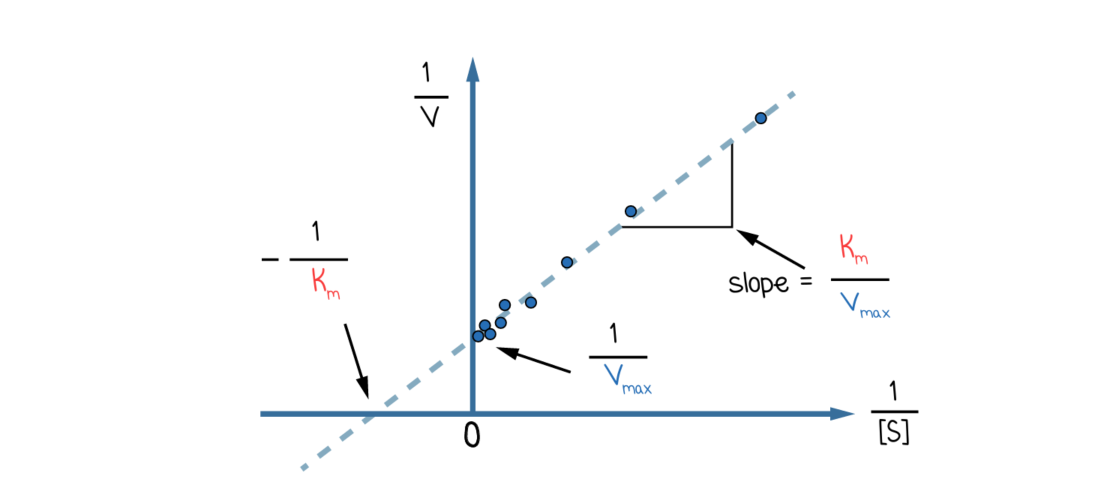
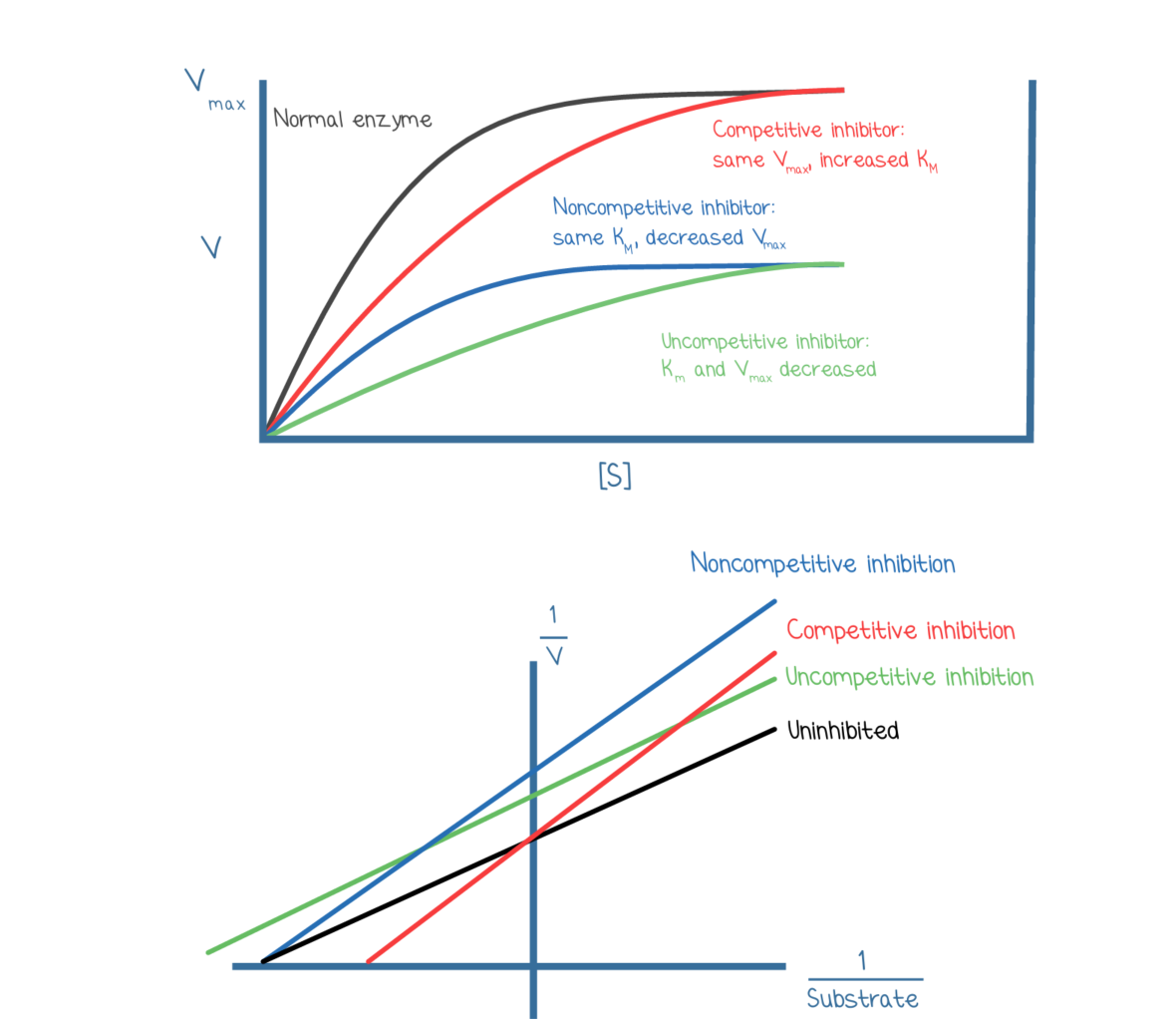
Lineweaver-Burk plots
Lineweaver-Burke plots are the double-reciprocal of Michaelis-Menten plots.
It is a linear 1/v vs 1/s graph with its slop being km/vmax
X intercept: -1/Km
Y intercept: 1/Vmax
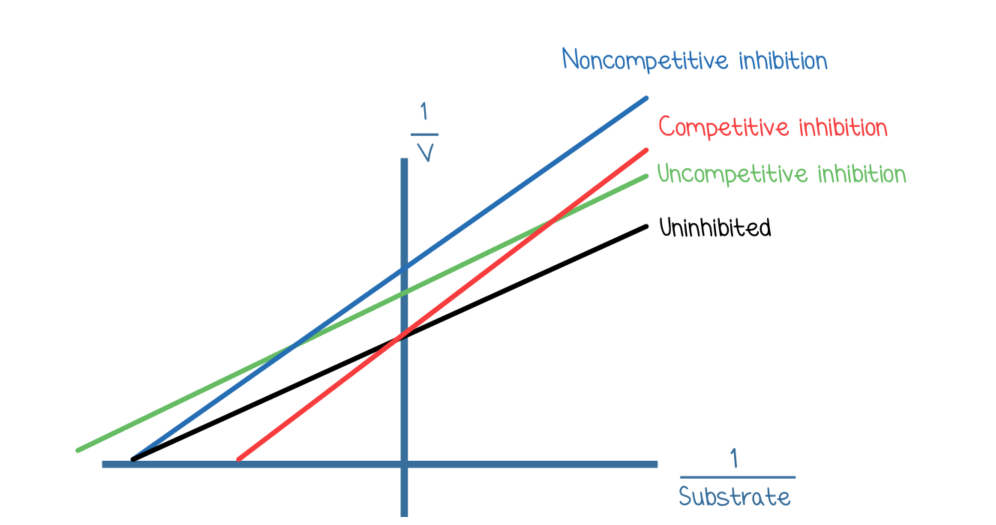
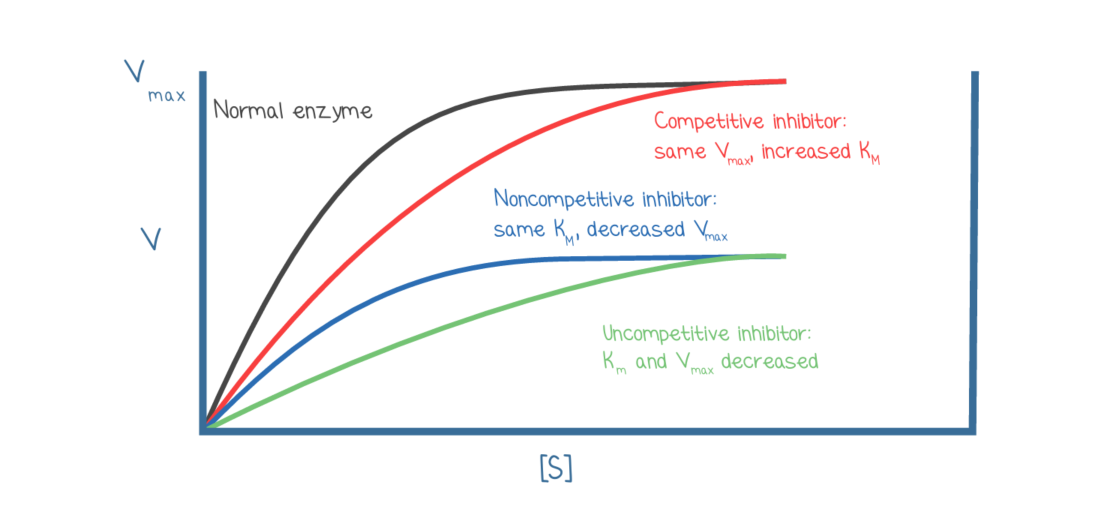
Effects of inhibitions on enzyme kinetics:
Competitive: increases Km, no affect on Vmax
binds to active site
moves x-intercept closer to origin on Lineweaver-Burk plots
Noncompetitive: no affect on Km, decreases Vmax
binds allosterically
moves y-intercept away from origin
Uncompetitive: decreases both Km and Vmax
Allosterically or near E-S complex
moves both the x-intercept and y-intercept away from the origin
Mixed: either decrease or increase Km and decreases Vmax
can either move the x-intercept away or towards the origin and make the y intercept away from the origin
if the mixed inhibitor ends up binding more readily to the enzyme- Km is higher
if the mixed inhibitor binds more readily to the enzyme-substrate complex- Km is lower
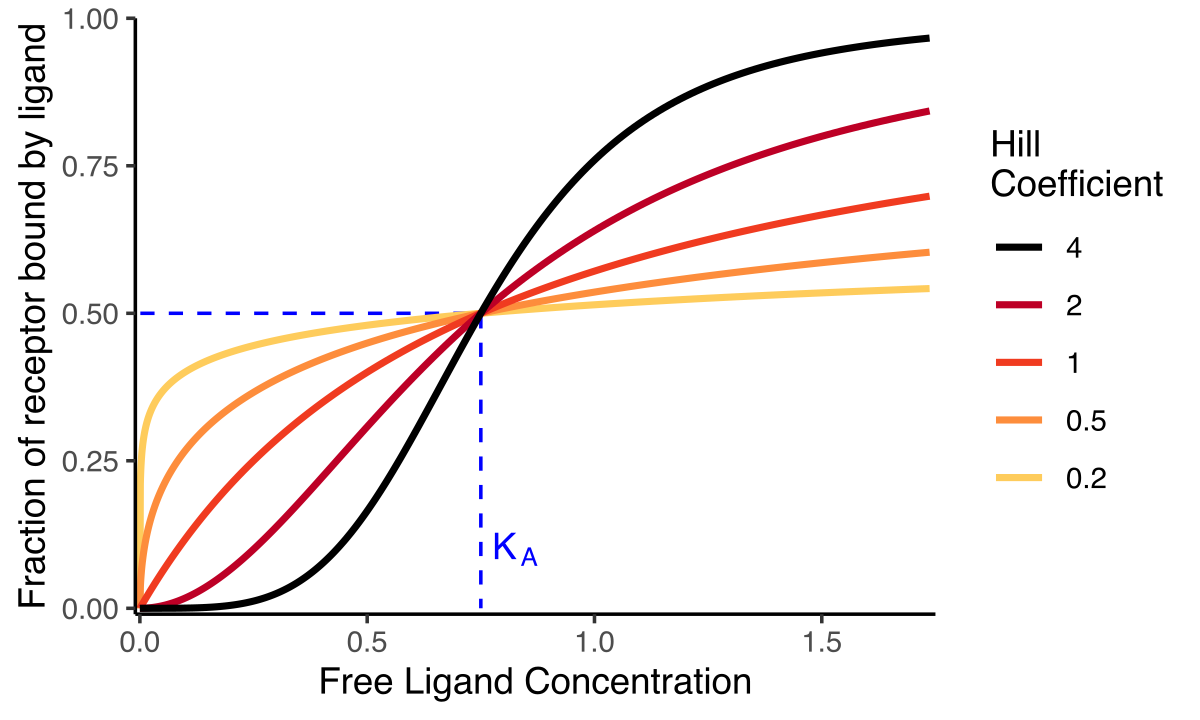
Cooperativity
Some enzymes or proteins can house more than one substrate at a time.
Sometimes, substrate binding changes substrate affinity (cooperativity)
Positive cooperative binding: substrate binding increases affinity for subsequent substrates
present in hemoglobin
results in a sigmoidal graph shape
Negative cooperative binding: substrate decreases affinity for subsequent substrates
results in hyperbolic graph shape
Non-cooperative binding: substrate binding does not affect affinity for subsequent substrates
present in myoglobin
hyperbolic graph shape
Hill Coefficients measure cooperativity:
n<1: negative cooperativity
n=1 no cooperativity (normal MM kinetics)
n>1: positive cooperability