Cell Bio- Ch. 7 Gene Expression
From RNA to DNA
Regulated information transfer links DNA sequences to cell functions
. DNA contains all info necessary for cell functions but does not perform any functions itself
Protein abundance can be controlled via mRNA abundance
. A gene that is highly transcribed and produces many mRNA transcripts leads to a greater amount of protein
.Different genes in the same cell and same gene in different cells can be expressed at different levels.
expression levels aren’t fixed, changing circumstances can change regulations
Structural differences between DNA and RNA
. RNA has ribose
ribose has a hydroxyl at c2, deoxyribose doesn’t
leads to important structural differences
.Nitrogenous bases
DNA uses thymine, RNA uses Uracil
thymine replaced uracil to make recognition of cytosine deamination easier
Uracil-DNA glycoslyase recognizes uracil in DNA and removes it.
Uracil pairs with adenine, just like thymine
Intramolecular Hydrogen bonding allows RNA to form folded structures
. RNA has not complementary partner
.Self-compliments, forms complex structure
. unconventional base-pairing causes folding and provides stabilizations
Transcription uses RNA as a template
. Synthesis from Template Strand
Matches Coding Strand
.RNA synthesis catalyzed by RNA polymerase
Multiple RNA transcripts can be transcribed in a series
. once one RNA polymerase leaves transcription site, another can move in
.bottlebrush appearance
.translation of a bacterial mRNA can begin before transcription is complete!
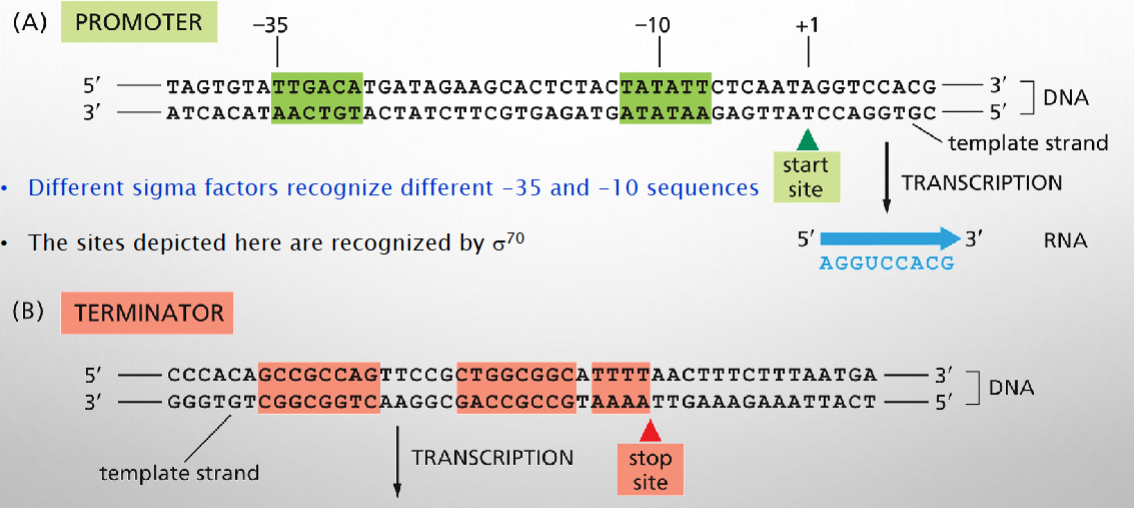
DNA sequences define transcription start+stop sites
. Ever gene has conserved sequences near beginning and end
Promoters are regulatory sequences just before start sites
.DNA regions that are found to have these sites with transferable sequences in between them are called open reading frames (ORFs)
.Sigma factors recruit RNA polymerase during bacterial transcription
Bacterial transcription consensus sequences
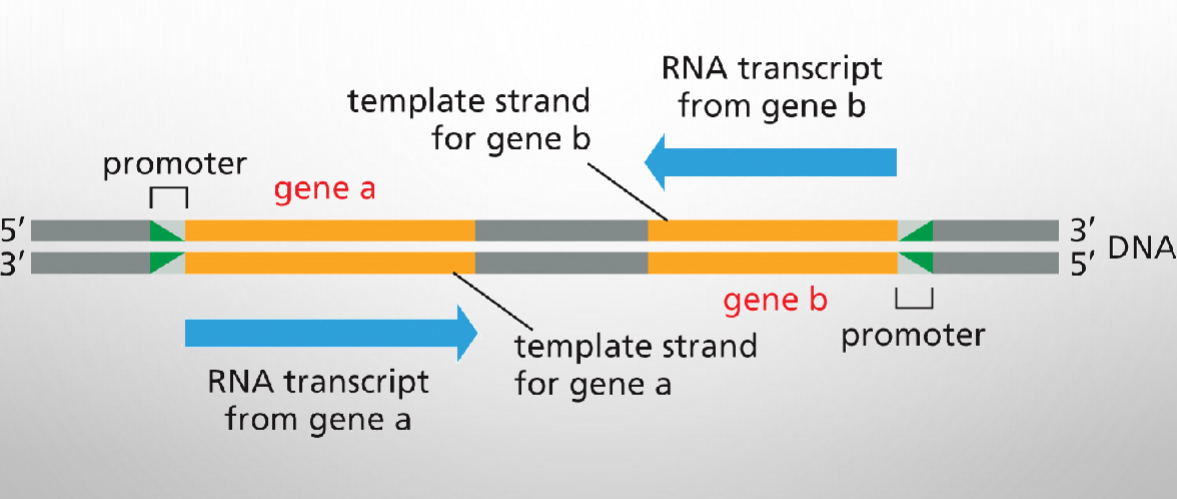
Either DNA strand can function as a template for Transcription
Eukaryotic transcription has more steps than prokaryotes
. Bacterial mRNAs have short life span
. Eukaryote mRNA needs to survive longer, so it needs to mature
Removal of non-coding introns
5’ cap addition
Poly-A tail addition
Different eukaryotic RNA polymerases produce different RNA types
. Which polymerase is the most active?: Polymerase 1
Consensus sequences proximal to eukaryotic transcription start site
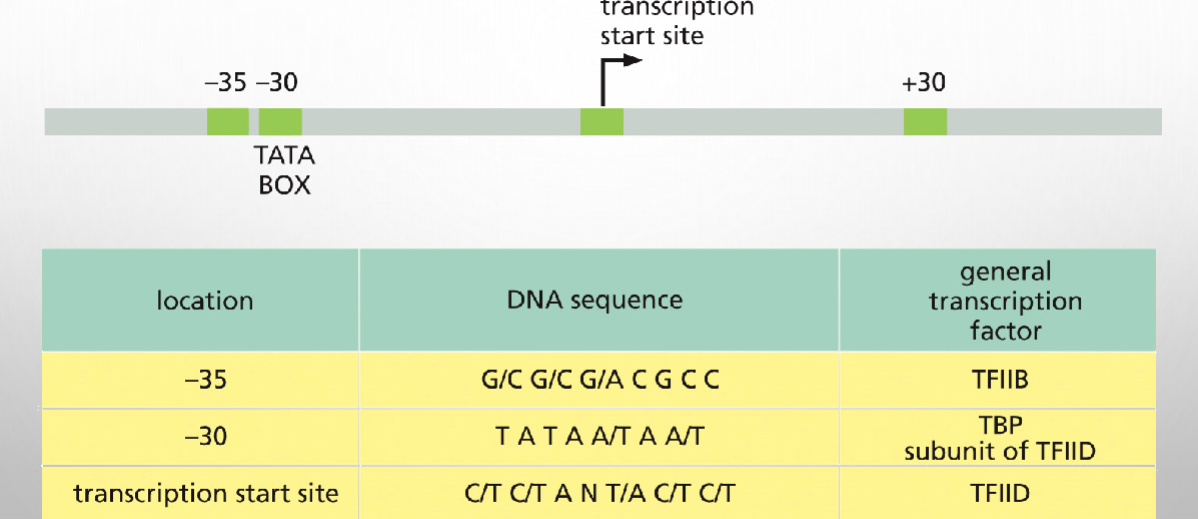
General transcription factors assemble into transcription initiation complex
RNA polymerase begins transcribing once it’s tail has been phosphorylated
.One of the final events of transcription initiation is phosphorylation tail domain
. Tail phosphorylation released RNA polymerase from initiation complex.
. Phosphorylation also generates docking sites for RNA processing enzymes including:
capping factors
splicing factors
polyadenylation factors
The docked factors ride along with RNA polymerase in close proximity to Nascent RNA
Eukaryotic mRNA is modified prior to nuclear export
.The 5’ cap is a methylated GTP
cap added by capping factors
linkage is an unconventional 5’-5’ triphosphate bridge
.Polyadenylation is the process of adding a string of adenines to the 3’ end of mRNA
poly-A tail plays a role in the initiation of translation
Poly-A tail also increases mRNA life span
Immature eukaryotic mRNA contains both coding and non-coding sequences
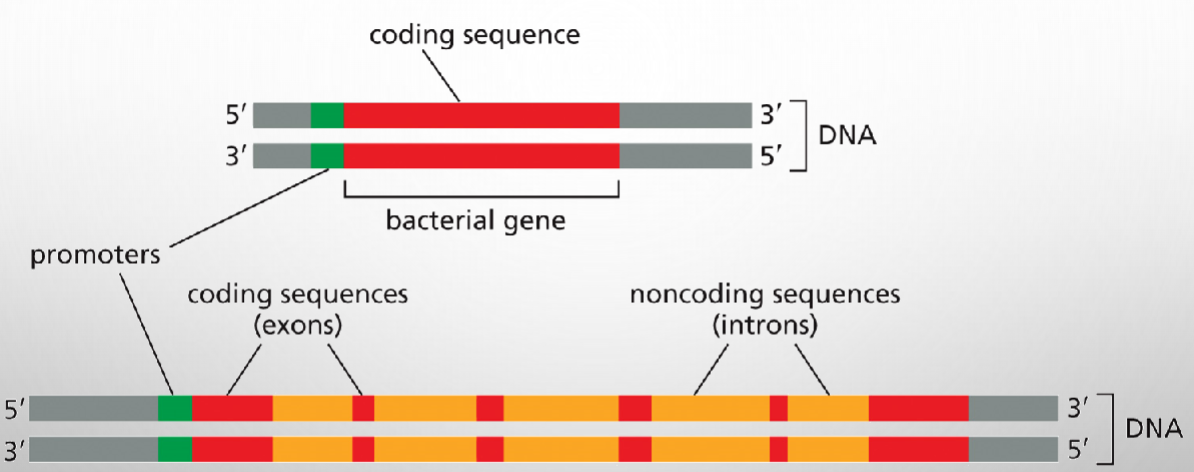
Intron size and number are variable
.Introns are identified by the exons on either side-the first intron of a gene would have the designations intron 1-2 because it is between exons 1 and 2
Consensus sequences mark introns and control their removal
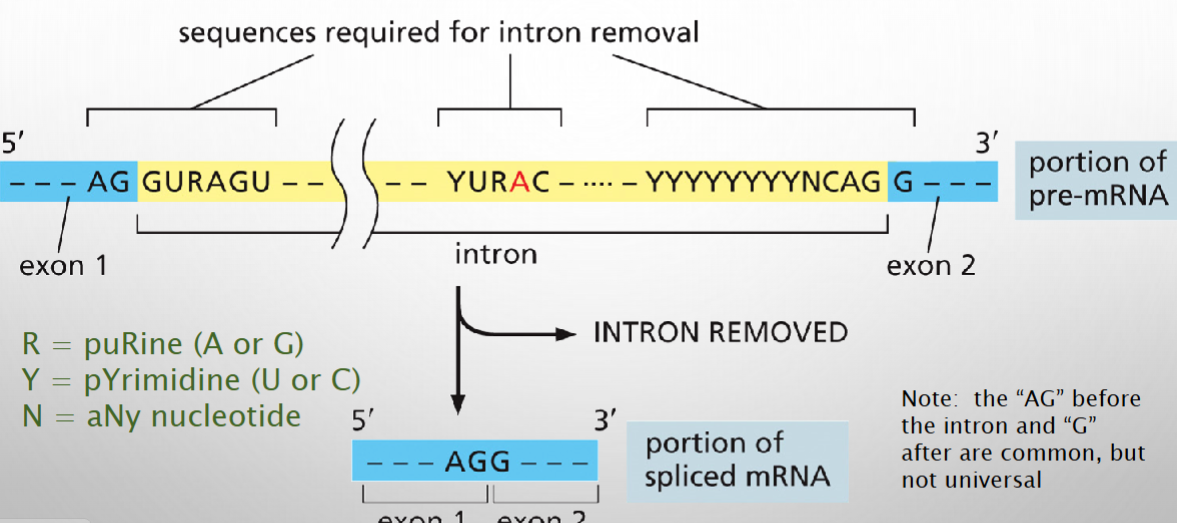
Splicing Removes an Intron and joins adjacent exons
.Components of spliceosome bring 5’ intron end in close proximity to the conserved adenine residue of the brand point
.The 2’ hydroxyl forms a bond with the 5’ intron end, detaching it from the 5’ splice donor and forming a lariat structure.
Small nuclear RNAs (snRNAs) and proteins catalyze intron removal
. Are ribozymes-non coding catalytic RNA
.snRNAs partner with proteins to form small nuclear ribonuclear proteins (snRNPs)
.Introns are initally recognized by U1 and U2 through base complementarity
.U6 double checks the 5’ intron end, displaces U1 and pairs with U2
.Interactions with different snNRPs and ATP hydrolysis forms the active site that catalyzes 5’ intron end bond transfer from the 5’ splice donor to the branch point.
.After lariat removal and splicing of exons, exon junction spanning proteins marks splicing as complete
Exon spanning proteins promote nuclear export and regulate translation
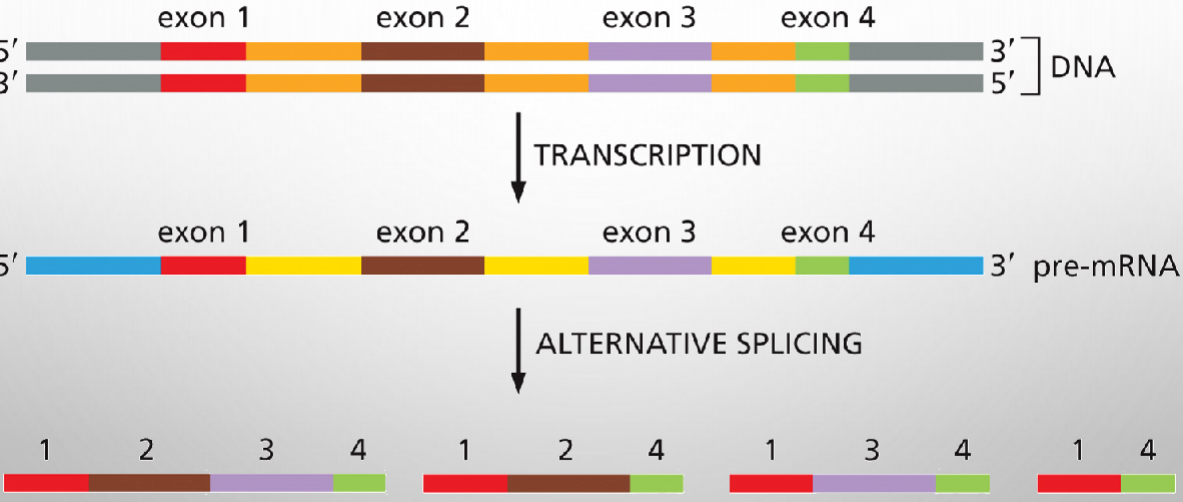
Alternative Splicing produces different mRNAs from same primary transcript
Mature eukaryotic mRNA is exported to the cytosol where it is translated
.After processing, mRNA is exported from the nucleus via nuclear pore complexes
.Export is enhanced by association of cap-binding, poly-A binding.
From RNA to Protein
RNA is translated into protein via 3-nucleotide “words”
.Codons
Start: AUG
Stop:UAA,UAG,UGA
. Relationship between codon sequence and final protein is called the genetic code.
code is degenerate, most amino acids are specified by more than one codon
The Genetic code
.Wheel model
Emphasizes the wobble of the third nucleotide
.The second nucleotide correlated well with the chemical nature of the specified amino acid
-U- specifies all hydrophobic amino acids. Hydrophilic acids represented by -A-
RNA reading frame dictated polypeptide amino acid composition
. Start point of translation is essential to producing correct polypeptide
.AUG sets reading frame for an mRNA sequence.
.Deletion of nucleotides can cause frameshift mutations
tRNA molecules “read” the genetic code
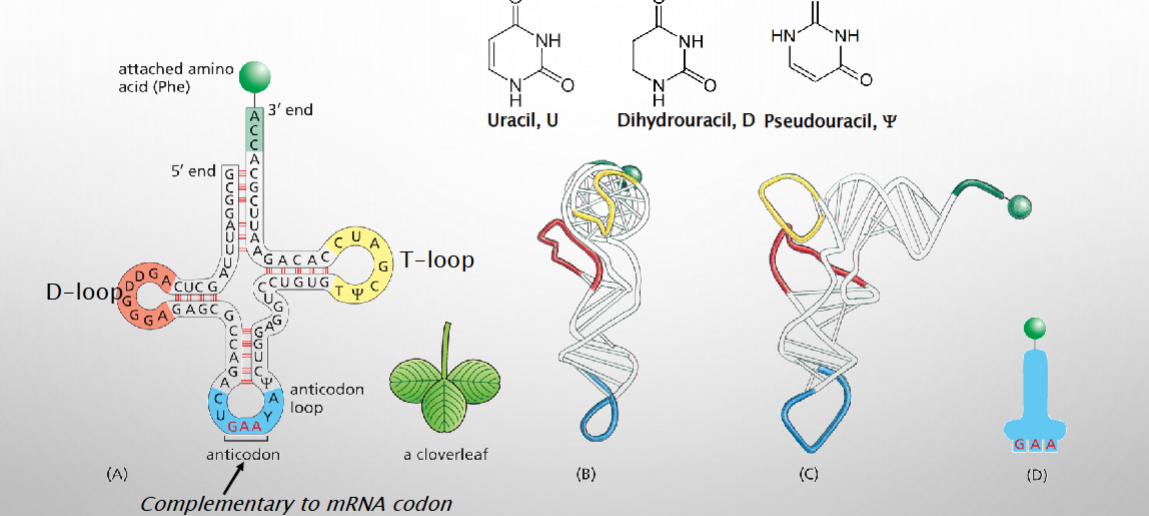
Aminoacyl-tRNA synthetases charge tRNAs with amino acid
.tRNAs must be loaded with amino acid to deliver one
. There is one synthetase for every amino acid.
each one can recognize more than on tRNA
Recognition relies on anticodon sequence and 3-D structure of tRNA, which varies according to carried animo acid.
.tRNA charging requires power from ATP hydrolysis
Both aminoacyl-tRNA synthetases and tRNA are required to translate mRNA
.An amino acid is linked to the 3’ end
mRNA is decoded on ribosomes
. Translation occurs on ribosomes,complexes of RNA and proteins
.Free Ribosome
Ribosomes have 3 tRNA binding sites and one mRNA site
. A site-aminoacyl
.P site- Peptidyl
.E Site- exit
Steps of translation:
aminoacyl tRNA entry
Peptide
Large subunit translocation, tRNA repositioning
Small subunit translocation, tRNA spent ejected
Eukaryotic translation begins with recognition of a START codon
. Initiator tRNA forms a complex with the small ribosomal subunit and translation initiation factors prior to binding mRNA.
. Sequences the 5’ untranslated region, recruits the initiation complex, which slides down the mRNA until it encounters a start codon.
. Complex arrests at AUG
some additional consensus sequences can emphasize the start codon
assisting sequences and the start codon are a Kozak sequence
Shine-Dalgarno sequence guides bacterial ribosomes
The translation initiation complex recruits the large ribosomal subunit
. recognition of START codon releases initiation factors and recruitment of large ribosomal subunit
.Once the ribosome is assembled with the initiator tRNA in the P site, translation can begin.
Bacterial mRNAs can contain multiple START sites and encode multiple proteins
. bacterial genes can by polycistronic, they can encode more than one protein
.Polycistronic mRNAs can have multiple ribosome binding sites
since bacterial ribosomes bind anywhere, each cistron must have it’s own START codon and ribosome binding site
individual cistrons can be translated independently of each other.
Termination of translation: STOP codons are recognized by release factors
A polysome translates multiple proteins from same RNA simultaneously
Some proteins require covalent modifications and binding partners to become functional.
. Nascent polypeptides can begin folding as they are translated
.many folded proteins are inactive until bound to cofactors or covalently modified