DNA and Protein Synthesis
Key points:
- DNA replication is semiconservative. Each strand in the double helix acts as a template for synthesis of a new, complementary strand.
- New DNA is made by enzymes called DNA polymerases, which require a template and a primer (starter) and synthesize DNA in the 5' to 3' direction.
- During DNA replication, one new strand (the leading strand) is made as a continuous piece. The other (the lagging strand) is made in small pieces.
- DNA replication requires other enzymes in addition to DNA polymerase, including DNA primase, DNA helicase, DNA ligase, and topoisomerase.
Introduction
DNA replication, or the copying of a cell's DNA, is no simple task! There are about 333 billion base pairs of DNA in your genome, all of which must be accurately copied when any one of your trillions of cells divides (1).
The basic mechanisms of DNA replication are similar across organisms. In this article, we'll focus on DNA replication as it takes place in the bacterium E. coli, but the mechanisms of replication are similar in humans and other eukaryotes.
Let's take a look at the proteins and enzymes that carry out replication, seeing how they work together to ensure accurate and complete replication of DNA.
The basic idea
<<DNA replication is semiconservative, meaning that each strand in the DNA double helix acts as a template for the synthesis of a new, complementary strand.<<
This process takes us from ^^one starting molecule to two "daughter" molecules^^, with each newly formed double helix containing one new and one old strand.
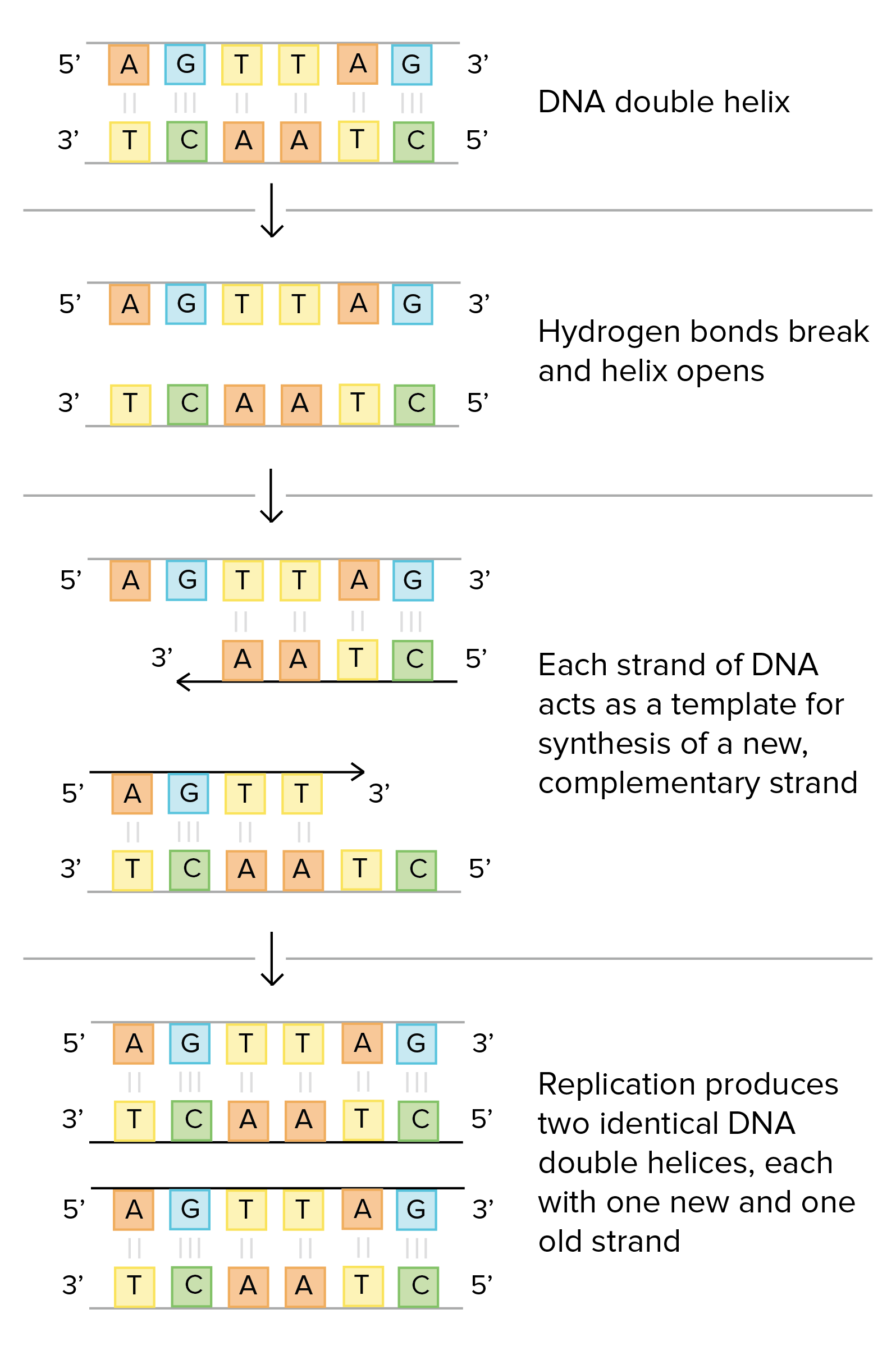
In a sense, that's all there is to DNA replication! But what's actually most interesting about this process is how it's carried out in a cell.
Cells need to copy their DNA very quickly, and with very few errors (or risk problems such as cancer). To do so, they use a variety of enzymes and proteins, which work together to make sure DNA replication is performed smoothly and accurately.
DNA polymerase
One of the key molecules in DNA replication is the enzyme DNA polymerase. DNA polymerases are responsible for synthesizing DNA: they add nucleotides one by one to the growing DNA chain, incorporating only those that are complementary to the template.
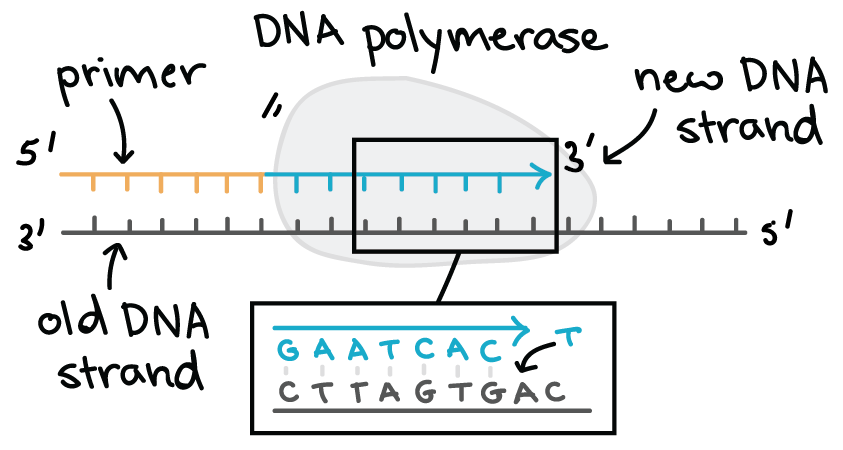
Here are some key features of DNA polymerases:
- They always need a template
- They can only add nucleotides to the 3' end of a DNA strand
- They can't start making a DNA chain from scratch, but require a pre-existing chain or short stretch of nucleotides called a primer
- They proofread, or check their work, removing the vast majority of "wrong" nucleotides that are accidentally added to the chain
The addition of nucleotides requires energy. This energy comes from the nucleotides themselves, which have three phosphates attached to them (much like the energy-carrying molecule ATP). When the bond between phosphates is broken, the energy released is used to form a bond between the incoming nucleotide and the growing chain.
[[See the polymerization reaction]](javascript:void(0))
In prokaryotes such as E. coli, there are two main DNA polymerases involved in DNA replication: DNA pol III (the major DNA-maker), and DNA pol I, which plays a crucial supporting role we'll examine later.
Starting DNA replication
How do DNA polymerases and other replication factors know where to begin? Replication always starts at specific locations on the DNA, which are called origins of replication and are recognized by their sequence.
E. coli, like most bacteria, has a single origin of replication on its chromosome. The origin is about 245245245 base pairs long and has mostly A/T base pairs (which are held together by fewer hydrogen bonds than G/C base pairs), making the DNA strands easier to separate.
Specialized proteins recognize the origin, bind to this site, and open up the DNA. As the DNA opens, two Y-shaped structures called replication forks are formed, together making up what's called a replication bubble. The replication forks will move in opposite directions as replication proceeds.
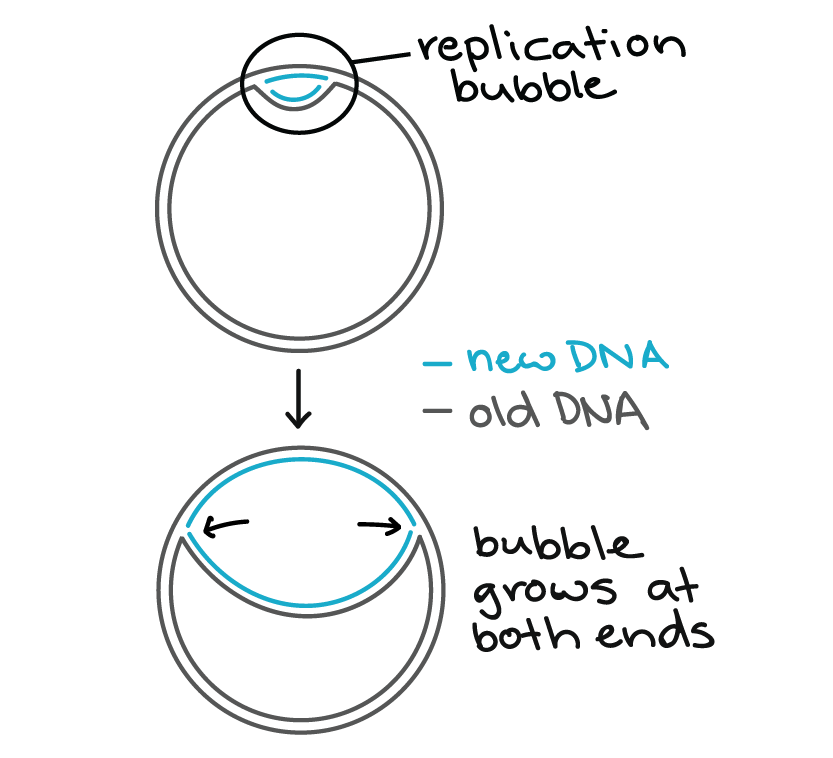
How does replication actually get going at the forks? Helicase is the first replication enzyme to load on at the origin of replication^33cubed. Helicase's job is to move the replication forks forward by "unwinding" the DNA (breaking the hydrogen bonds between the nitrogenous base pairs).
Proteins called single-strand binding proteins coat the separated strands of DNA near the replication fork, keeping them from coming back together into a double helix.
Primers and primase
DNA polymerases can only add nucleotides to the 3' end of an existing DNA strand. (They use the free -OH group found at the 3' end as a "hook," adding a nucleotide to this group in the polymerization reaction.) How, then, does DNA polymerase add the first nucleotide at a new replication fork?
Alone, it can't! The problem is solved with the help of an enzyme called primase. Primase makes an RNA primer, or short stretch of nucleic acid complementary to the template, that provides a 3' end for DNA polymerase to work on. A typical primer is about five to ten nucleotides long. The primer primes DNA synthesis, i.e., gets it started.
Once the RNA primer is in place, DNA polymerase "extends" it, adding nucleotides one by one to make a new DNA strand that's complementary to the template strand.
Leading and lagging strands
In E. coli, the DNA polymerase that handles most of the synthesis is DNA polymerase III. There are two molecules of DNA polymerase III at a replication fork, each of them hard at work on one of the two new DNA strands.
DNA polymerases can only make DNA in the 5' to 3' direction, and this poses a problem during replication. A DNA double helix is always anti-parallel; in other words, one strand runs in the 5' to 3' direction, while the other runs in the 3' to 5' direction. This makes it necessary for the two new strands, which are also antiparallel to their templates, to be made in slightly different ways.
One new strand, which runs 5' to 3' towards the replication fork, is the easy one. This strand is made continuously, because the DNA polymerase is moving in the same direction as the replication fork. This continuously synthesized strand is called the leading strand.
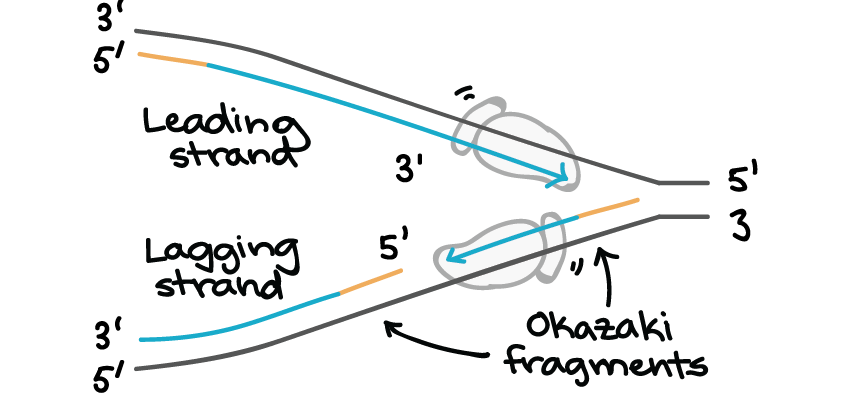
The other new strand, which runs 5' to 3' away from the fork, is trickier. This strand is made in fragments because, as the fork moves forward, the DNA polymerase (which is moving away from the fork) must come off and reattach on the newly exposed DNA. This tricky strand, which is made in fragments, is called the lagging strand.
The small fragments are called Okazaki fragments, named for the Japanese scientist who discovered them. The leading strand can be extended from one primer alone, whereas the lagging strand needs a new primer for each of the short Okazaki fragments.
The maintenance and cleanup crew
Some other proteins and enzymes, in addition the main ones above, are needed to keep DNA replication running smoothly. One is a protein called the sliding clamp, which holds DNA polymerase III molecules in place as they synthesize DNA. The sliding clamp is a ring-shaped protein and keeps the DNA polymerase of the lagging strand from floating off when it re-starts at a new Okazaki fragment^44start superscript, 4, end superscript.
Topoisomerase also plays an important maintenance role during DNA replication. This enzyme prevents the DNA double helix ahead of the replication fork from getting too tightly wound as the DNA is opened up. It acts by making temporary nicks in the helix to release the tension, then sealing the nicks to avoid permanent damage.
Finally, there is a little cleanup work to do if we want DNA that doesn't contain any RNA or gaps. The RNA primers are removed and replaced by DNA through the activity of DNA polymerase I, the other polymerase involved in replication. The nicks that remain after the primers are replaced get sealed by the enzyme DNA ligase.
Summary of DNA replication in E. coli
Let's zoom out and see how the enzymes and proteins involved in replication work together to synthesize new DNA.
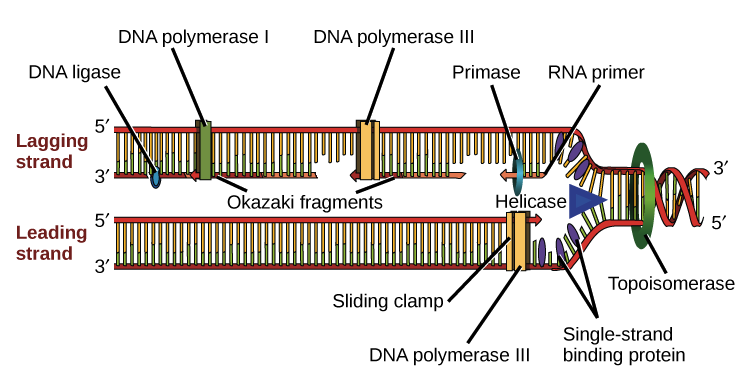
- Helicase opens up the DNA at the replication fork.
- Single-strand binding proteins coat the DNA around the replication fork to prevent rewinding of the DNA.
- Topoisomerase works at the region ahead of the replication fork to prevent supercoiling.
- Primase synthesizes RNA primers complementary to the DNA strand.
- DNA polymerase III extends the primers, adding on to the 3' end, to make the bulk of the new DNA.
- RNA primers are removed and replaced with DNA by DNA polymerase I.
- The gaps between DNA fragments are sealed by DNA ligase.
DNA replication in eukaryotes
The basics of DNA replication are similar between bacteria and eukaryotes such as humans, but there are also some differences:
- Eukaryotes usually have multiple linear chromosomes, each with multiple origins of replication. Humans can have up to 100,100,100, comma000000000 origins of replication^55start superscript, 5, end superscript!
- Most of the E. coli enzymes have counterparts in eukaryotic DNA replication, but a single enzyme in E. coli may be represented by multiple enzymes in eukaryotes. For instance, there are five human DNA polymerases with important roles in replication^55start superscript, 5, end superscript.
- Most eukaryotic chromosomes are linear. Because of the way the lagging strand is made, some DNA is lost from the ends of linear chromosomes (the telomeres) in each round of replication.