M1L5: Cell Cycle Controls and Radiation Induced Checkpoints
Cell cycle
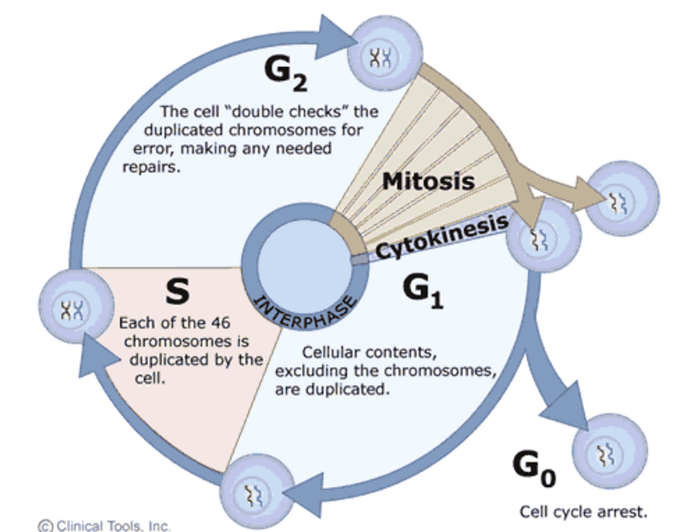
G1 - cellular contents are duplicated
S - DNA replication of all 46 chromosomes
G2 - cell ‘double checks’ the replicated chromosomes for errors and triggers repair if needed
Mitosis
Cytokinesis
G0 - cell cycle arrest
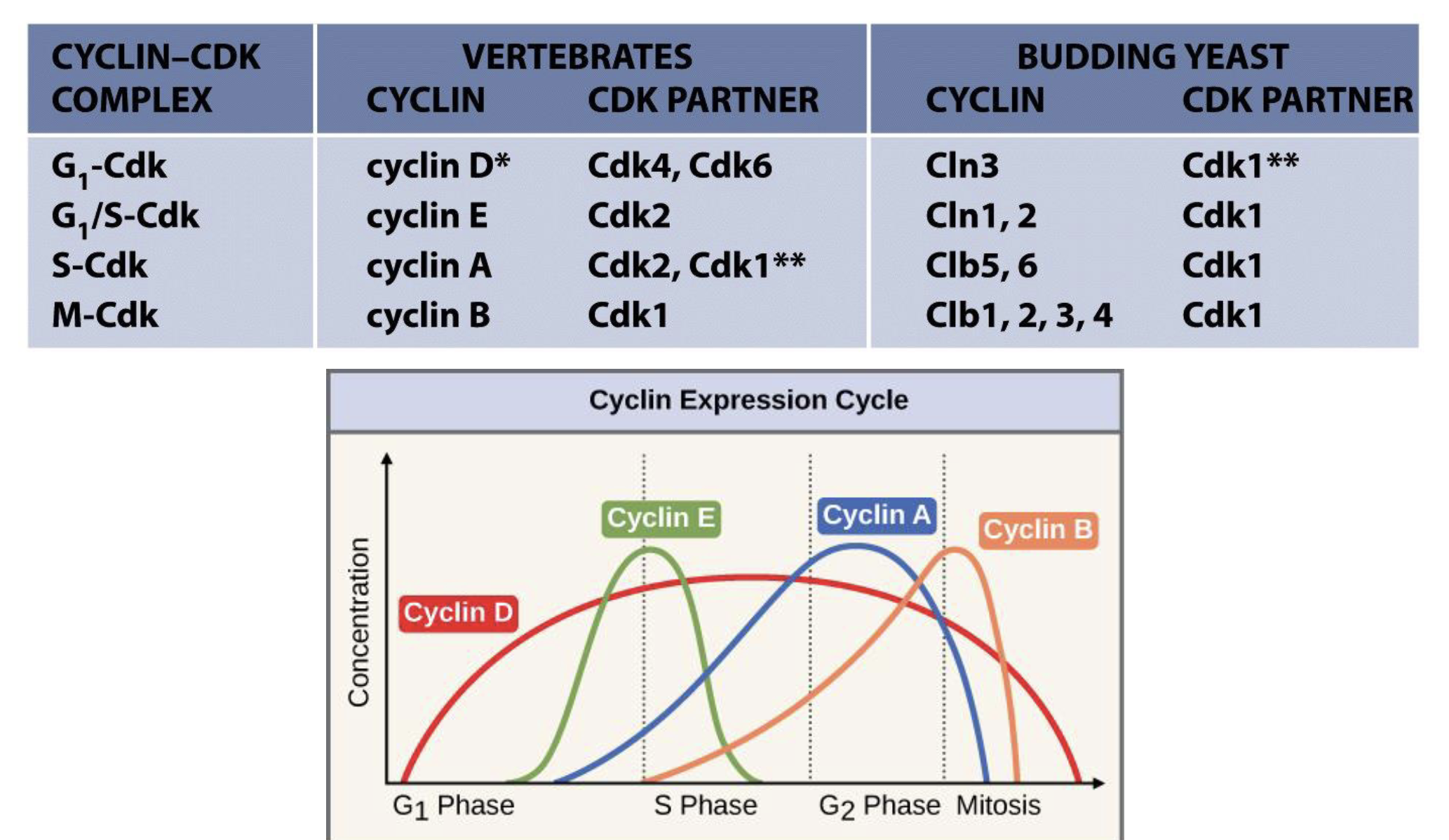
Control of cell cycle by cyclins and CDKs
Cell cycle control is controlled by cyclins/cyclin-dependent kinases (phosphorylation) and ubiquitination
There are many cyclins and Cdks in cells but not all regulate the cell cycle, the few that do are heterodimer kinases
Regulatory subunit - conc varies in cyclical fashion during cell cycle
Can be degraded by the ubiquitin system
Catalytic subunit (Cdk) - Ser/Thr kinase, always present but not always active
Cdks are active only with cyclin binding
Regulation of cyclin-Cdk activity
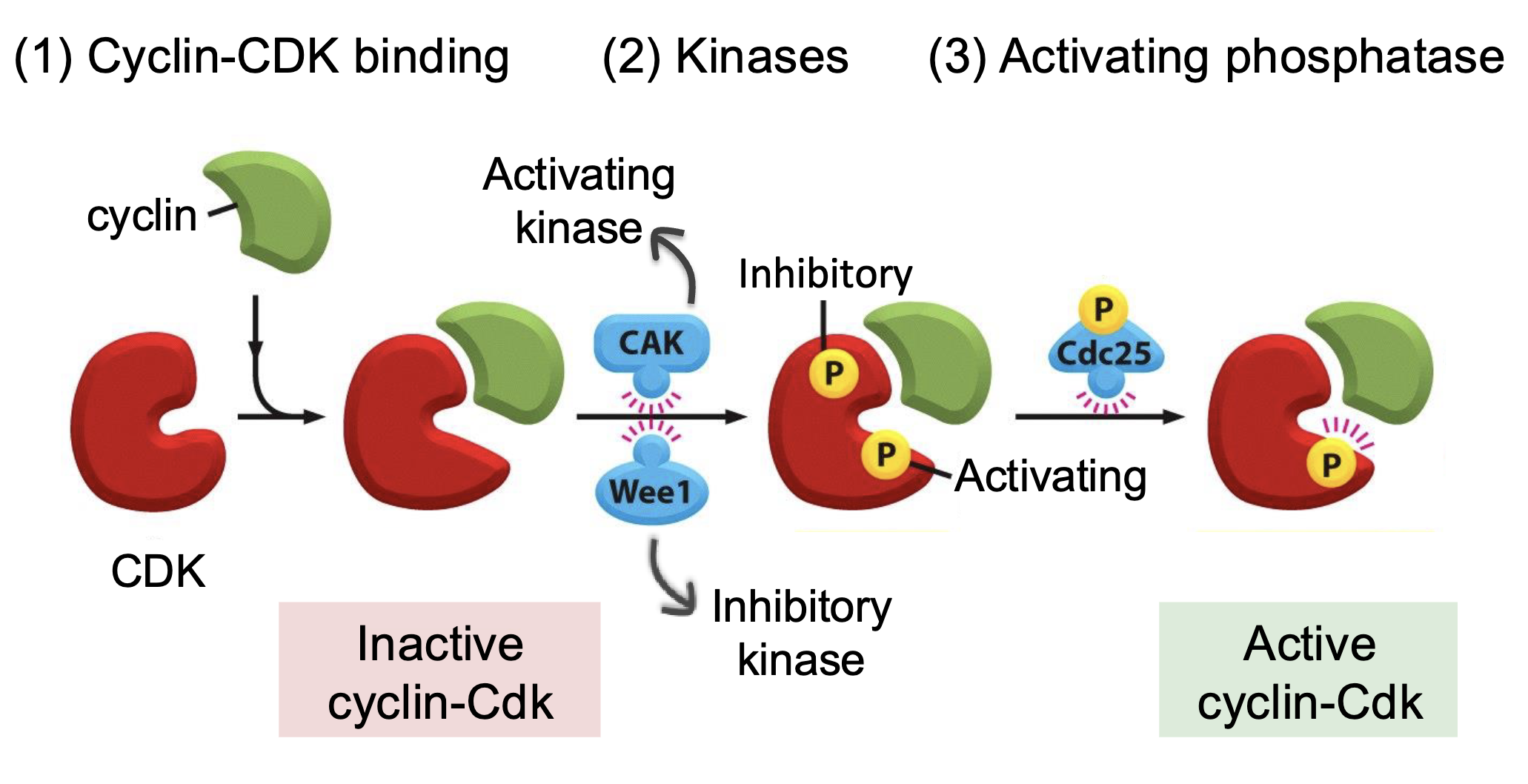
Cyclin-Cdk binding
Activating and inactivating kinases
Phosphorylation at specific sites can alter the protein conformation in a way that activates or inhibits its activity
Cak - activates Cdk
Wee1 - inhibits Cdk
Can be used by cancer cells to avoid mitotic catastrophe, trigger senescence/repair, secrete interleukins in this time to reprogram the TME
Activating phosphatase (Cdc25) - removes the phosphorylation from inhibitory kinase to activate the Cdk
Cdk inhibitory proteins inhibibit cell cycle progression by binding to Cdk
INK4 (p16INK4a, p15INK4b, p18INK4c, p19INK4d) - inhibits Cdk4 and Cdk6
Cip/Kip (p21Cip1, p27Kip1, p57Kip2) - inhibits Cdk2
Cell division checkpoints - mainly controlled by pRb (G1/S progression)
DNA damage induced checkpoints - mainly controlled by p53
Cell division checkpoints

Largely regulated by pRb
Start checkpoint (G1/S) /restriction point (R) - is environment favourable?
If conditions are favourable, and DNA is not damaged then cell commits to DNA replication, otherwise it can escape to G0 or trigger apoptosis
Execution of R point depends on expression of Rb
G2/M checkpoint - is all DNA replicated, is environment favourable?
If DNA has been correctly replicated and DNA is not damaged, cell proceeds to chromosome alignment on spindle in metaphase
Metaphase to anaphase transition/spindle assembly checkpoint - are all chromosomes attached to spindle?
If all chromosomes are correctly attached to spindle microtubules then cell proceeds to anaphase
No checkpoint in S phase - ‘artificial’ or ‘mild’ checkpoints only, mainly by DNA damage
Likely due to high fidelity of DNA polymerases in DNA synthesis
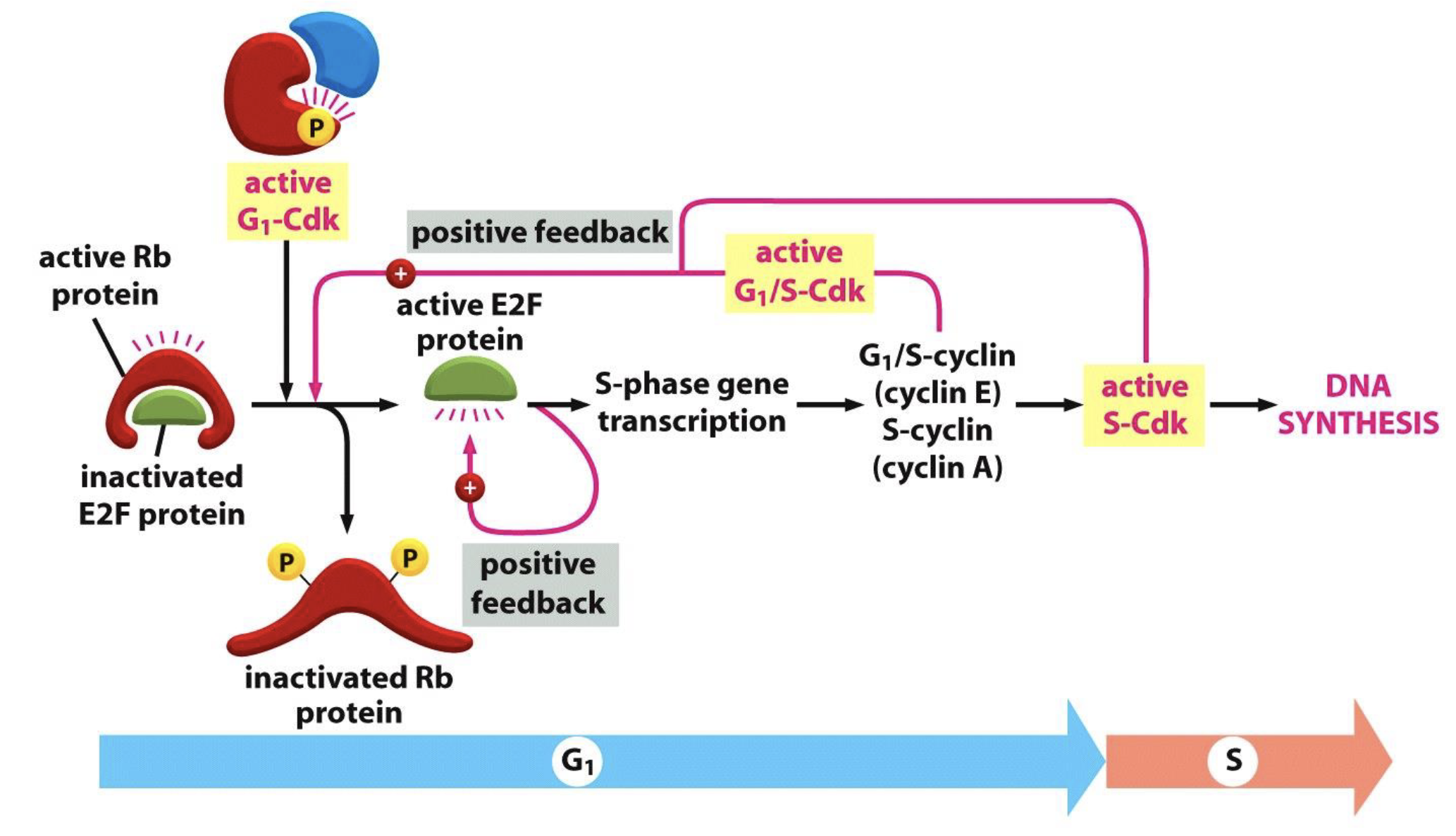
Control of S phase entry by pRb
Active Rb binds to E2F family of transcription factors to inactivate it, arresting the cell cycle in G1
G1-Cdk (cyclin D-Cdk4/6) phosphorylate Rb to inactivate it which releases E2F
Active E2F triggers transcription of S phase genes, including G1/S cyclin (cyclin E) and S-cyclin (cyclin A), and also itself (positive feedback)
Cyclin E and A production activates S-Cdk (cyclin A-Cdk2) and causes DNA synthesis
Cyclin E–Cdk2 (G₁/S-Cdk) and cyclin A-Cdk2 (S-Cdk) further phosphorylate Rb to fully inactivate it (positive feedback)
Mitogens and DNA damage compete to regulate cyclin:Cdk inhibitors balance and S phase entry
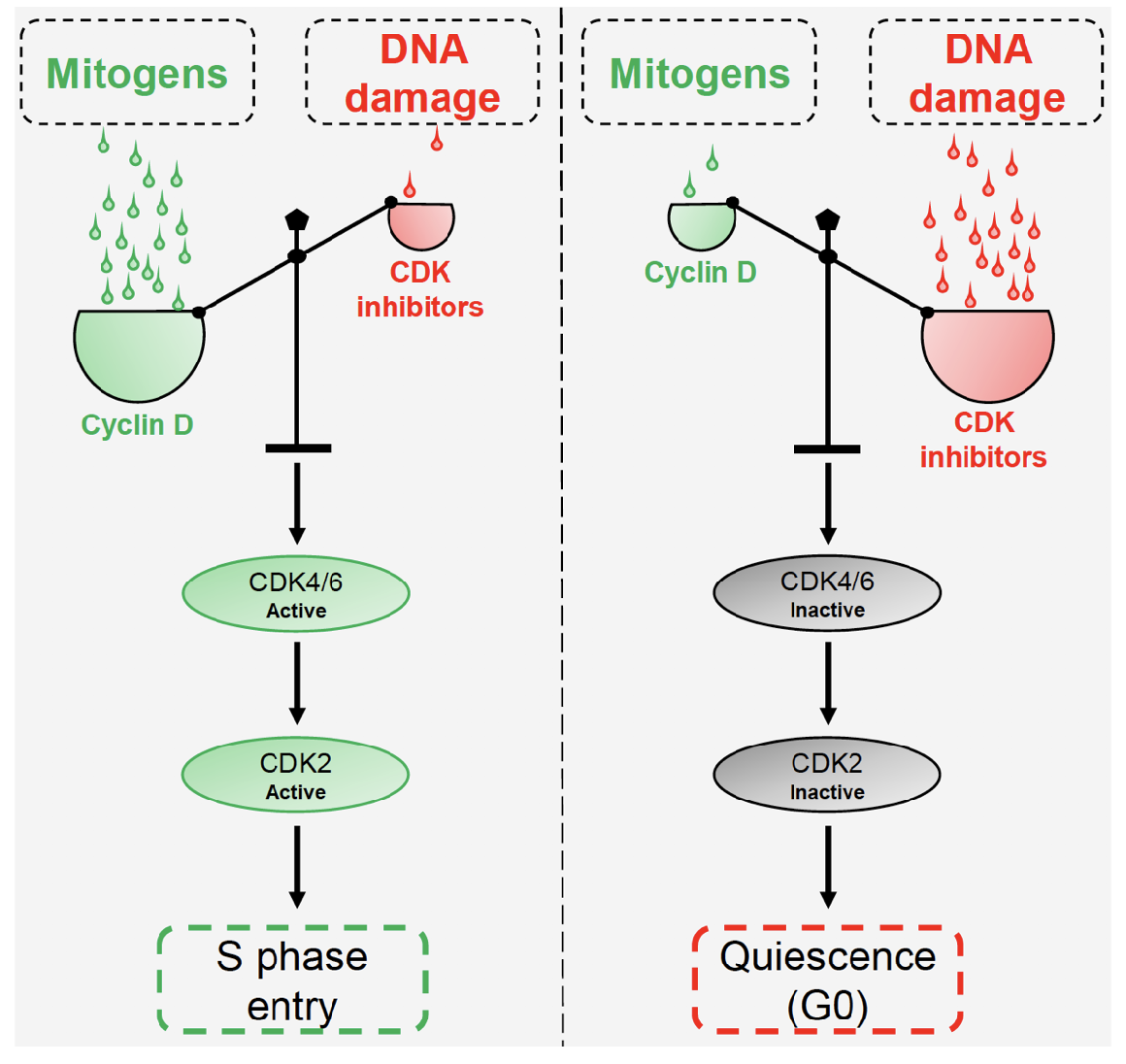
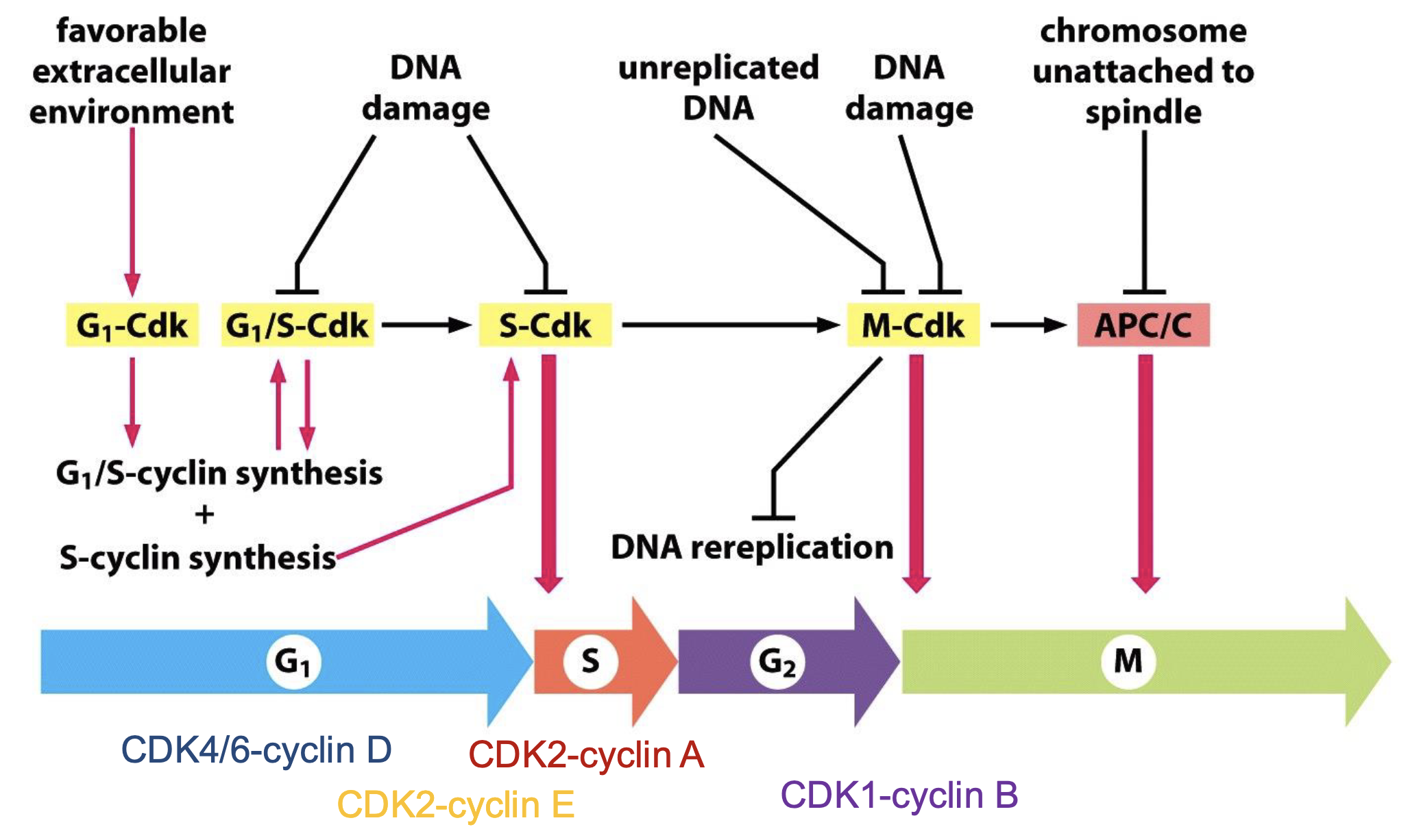
Overview of cell cycle control system
DDR (endogenous and exogenous)
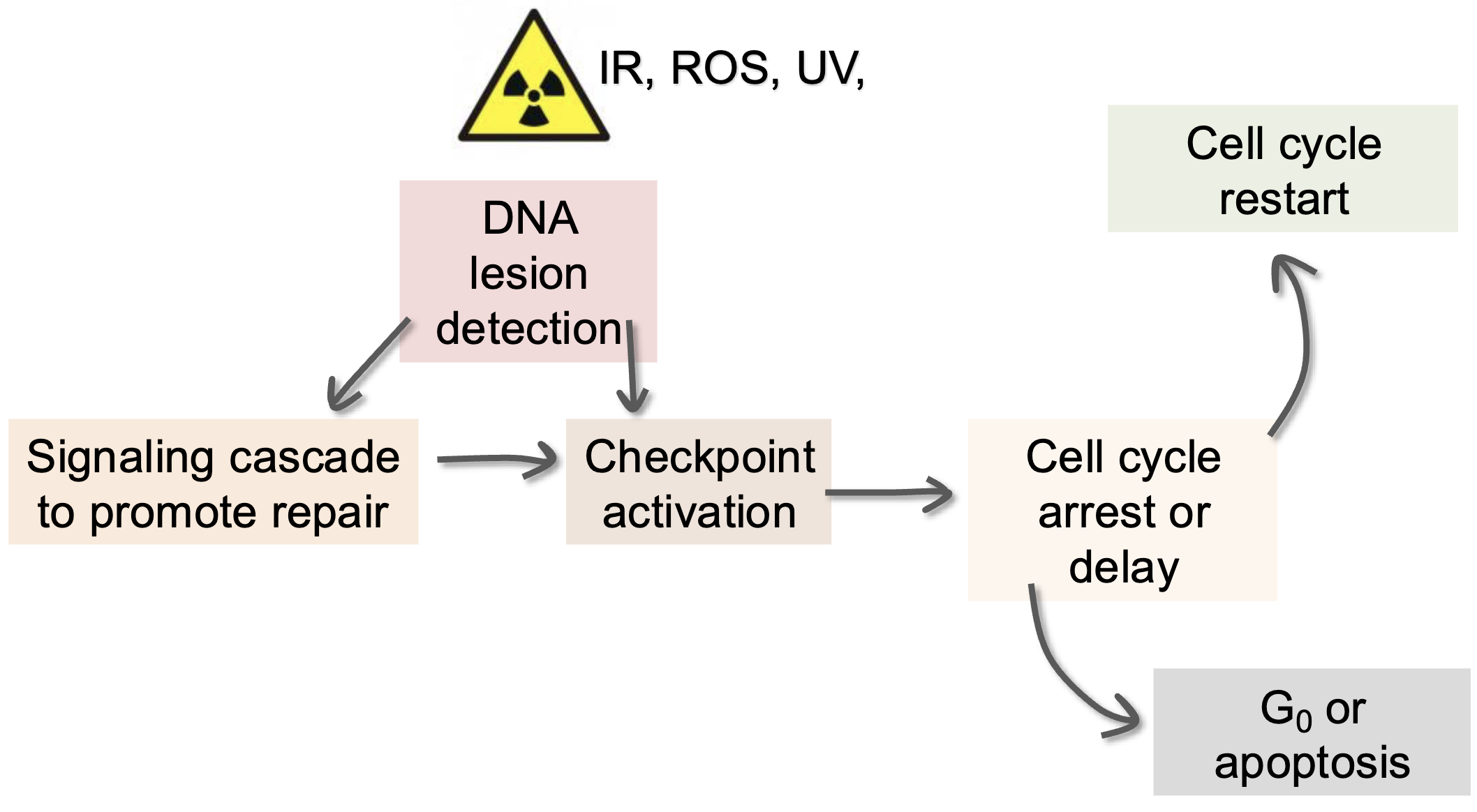
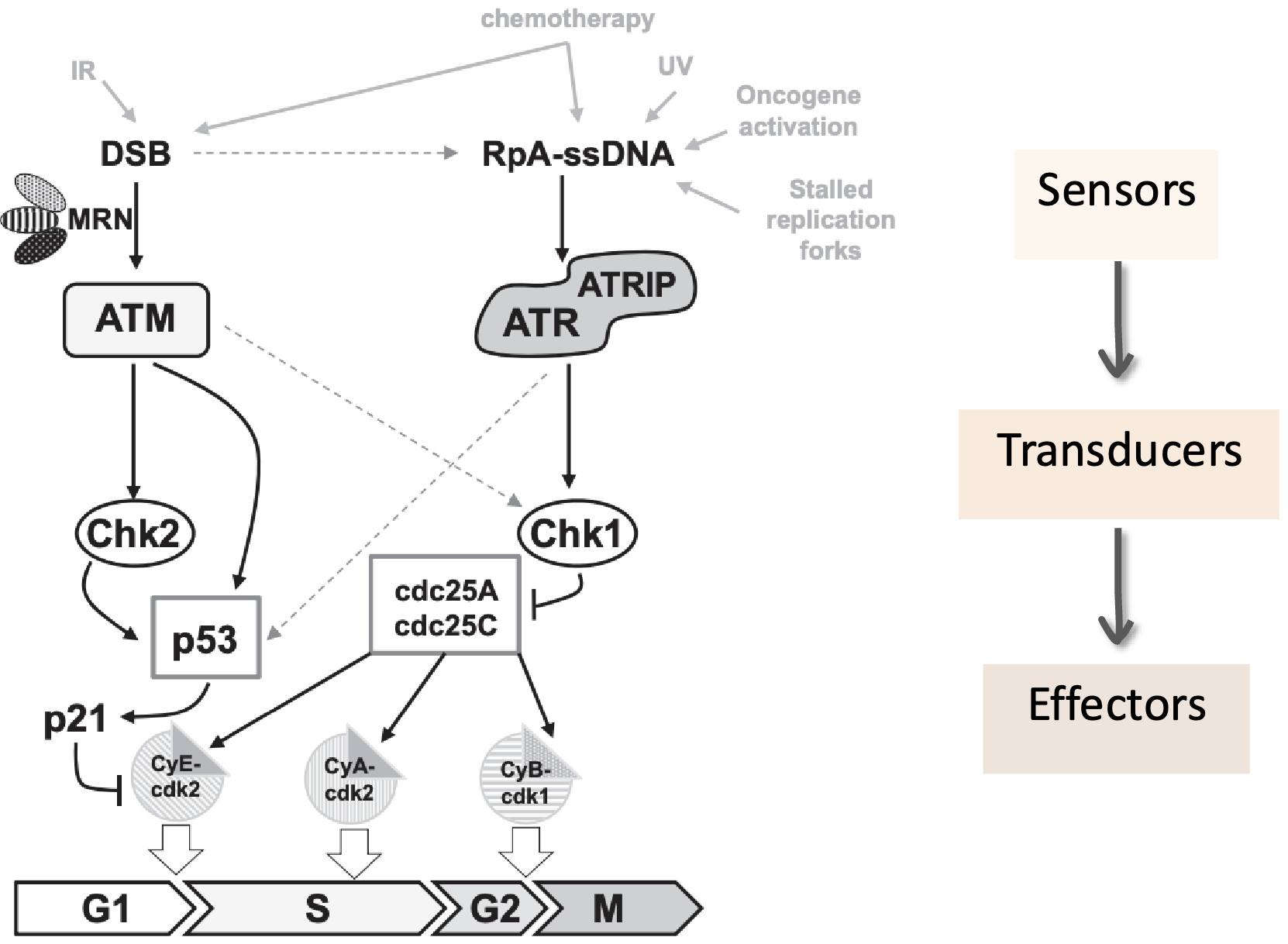
DNA-damage induced checkpoints
Depending on when DNA damage is detected
G1 checkpoint - leads to accumulation of cells in G1 or G1 arrest
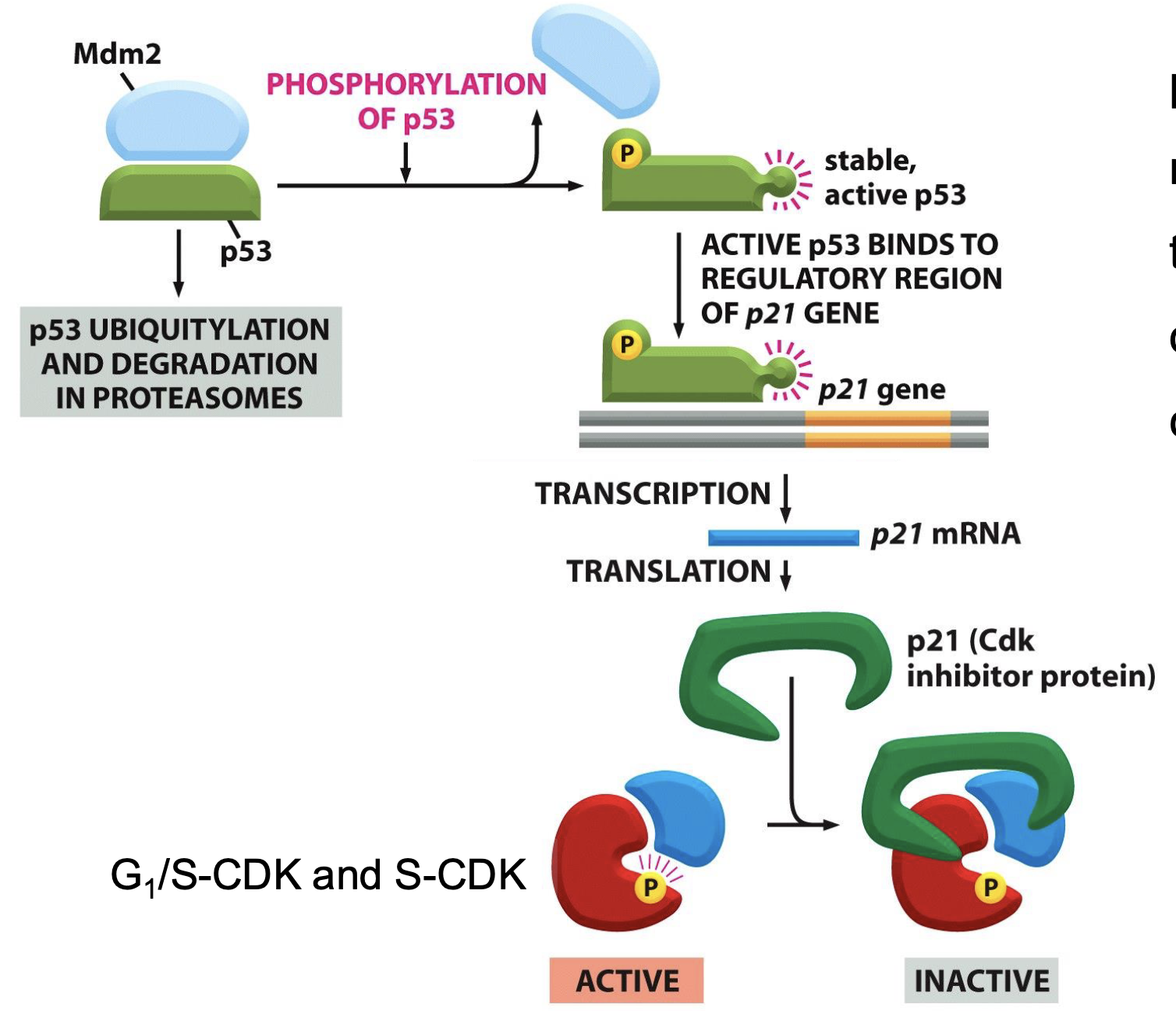
p53-p21 pathway (slow)
p53 complexes with the E3 ligase Mdm2 which shuttles p53 into the proteasome for degradation
p53 phosphorylation stabilises/activates p53
p53 binds to promoter region of p21 gene - triggering transcription and translation to produce p21 protein
p21 acts as a Cdk inhibitor (cyclin E/Cdk2 and cyclin D/Cdk4/6) - normally CycD/Cdk4/6 phosphorylates pRb to release E2F which promotes S phase gene transcription, including CycE/Cdk2 which further phosphorylates pRb (positive feedback)
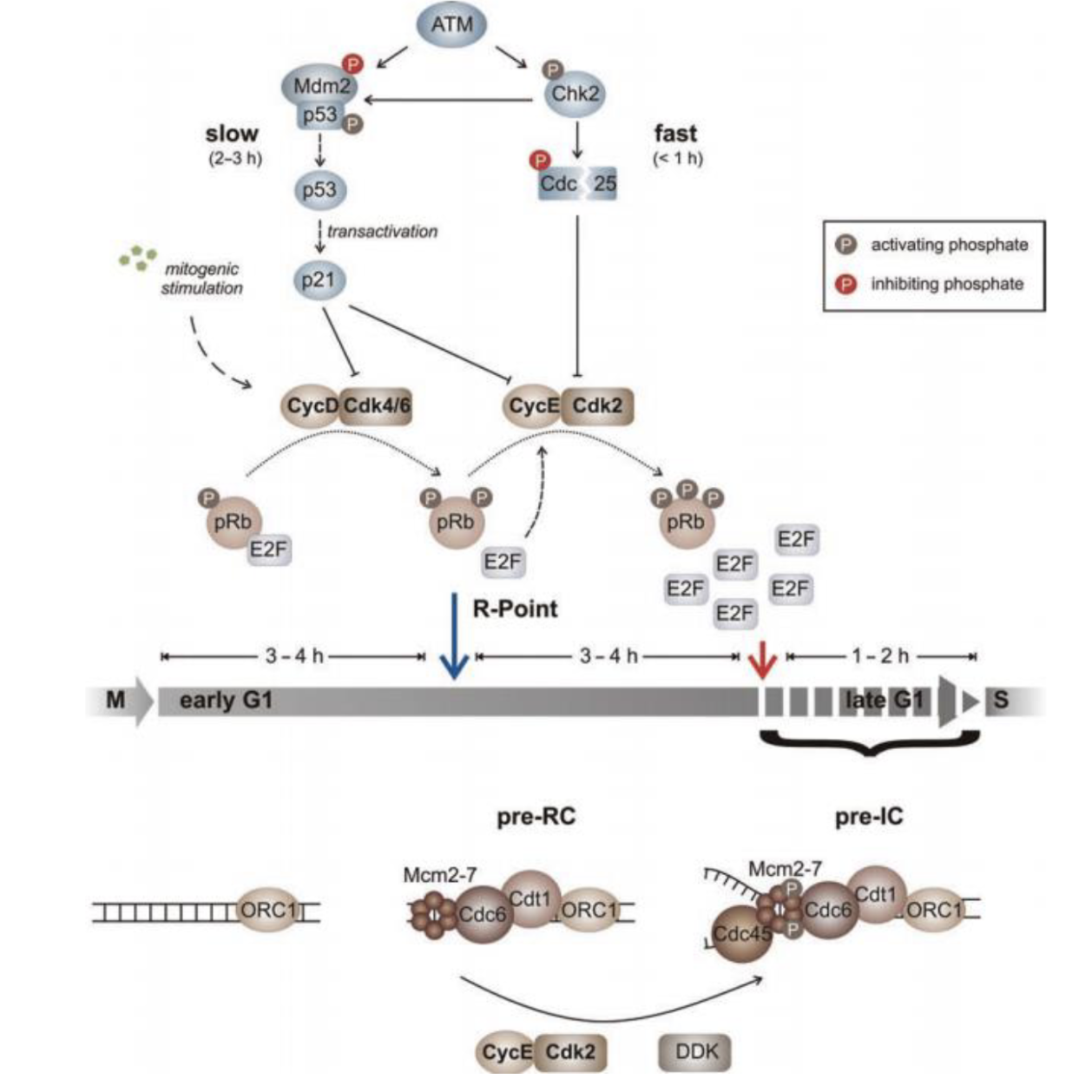
Chk2 pathway - ATM activates Chk2 by phosphorylation which activates the checkpoint by inhibiting Cdc25 which in turn inhibits CycE/Cdk2
Intra S checkpoint or replication checkpoint - slows or shuts down replication initiation and elongation
Fork dependent (eg. dNTP depletion, collision of fork with damaged DNA, fork stalling leading to ssDNA)
Fork stalling results in ssDNA which gets coated by RPA which recruits ATRIP which in turn recruits ATR
ATR phosphorylates CHK1 to activate the checkpoint by inhibiting Cdc25A/C
RAD51 and BRCA1/2 help stabilise the fork and chromatin remodelers
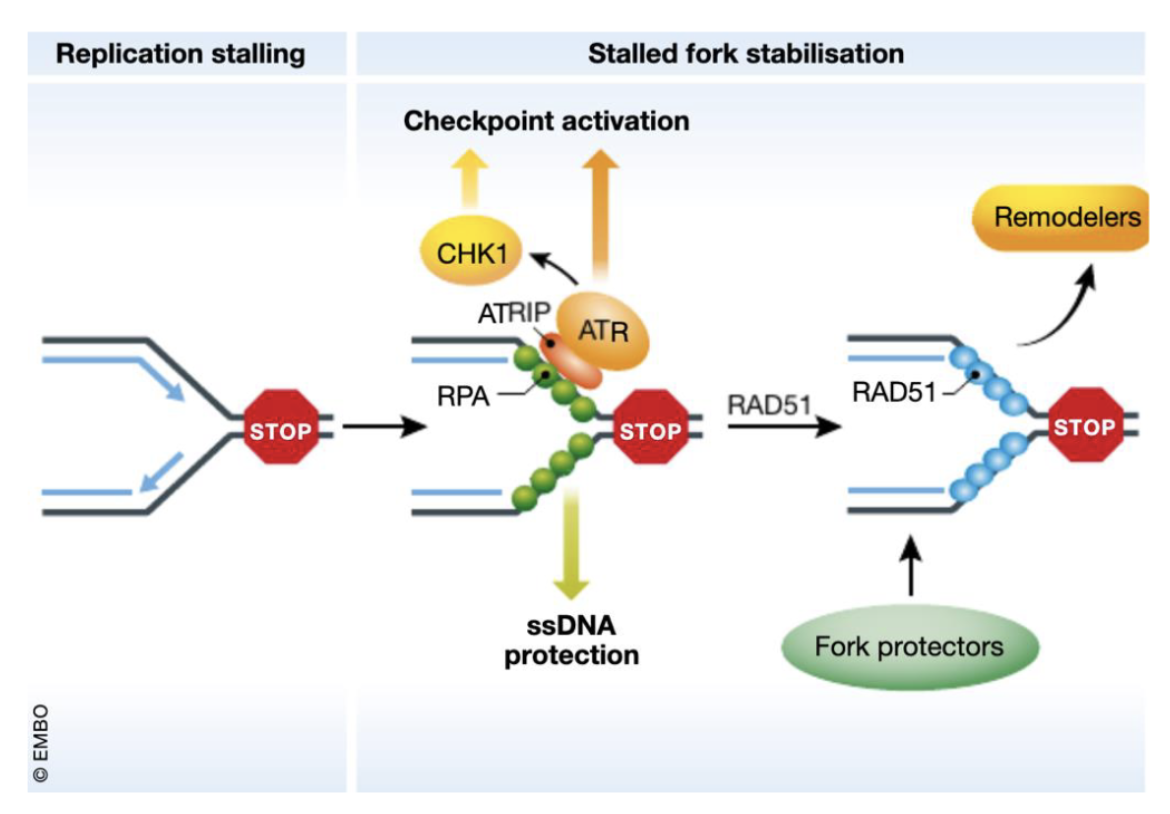
In the fork independent route (DSBs eg due to IR), structure-specific nucleases like MUS81 process the DNA ends and
DNA polymerase δ subunit POLD3 can help restart replication through break-induced replication (BIR)–like mechanisms
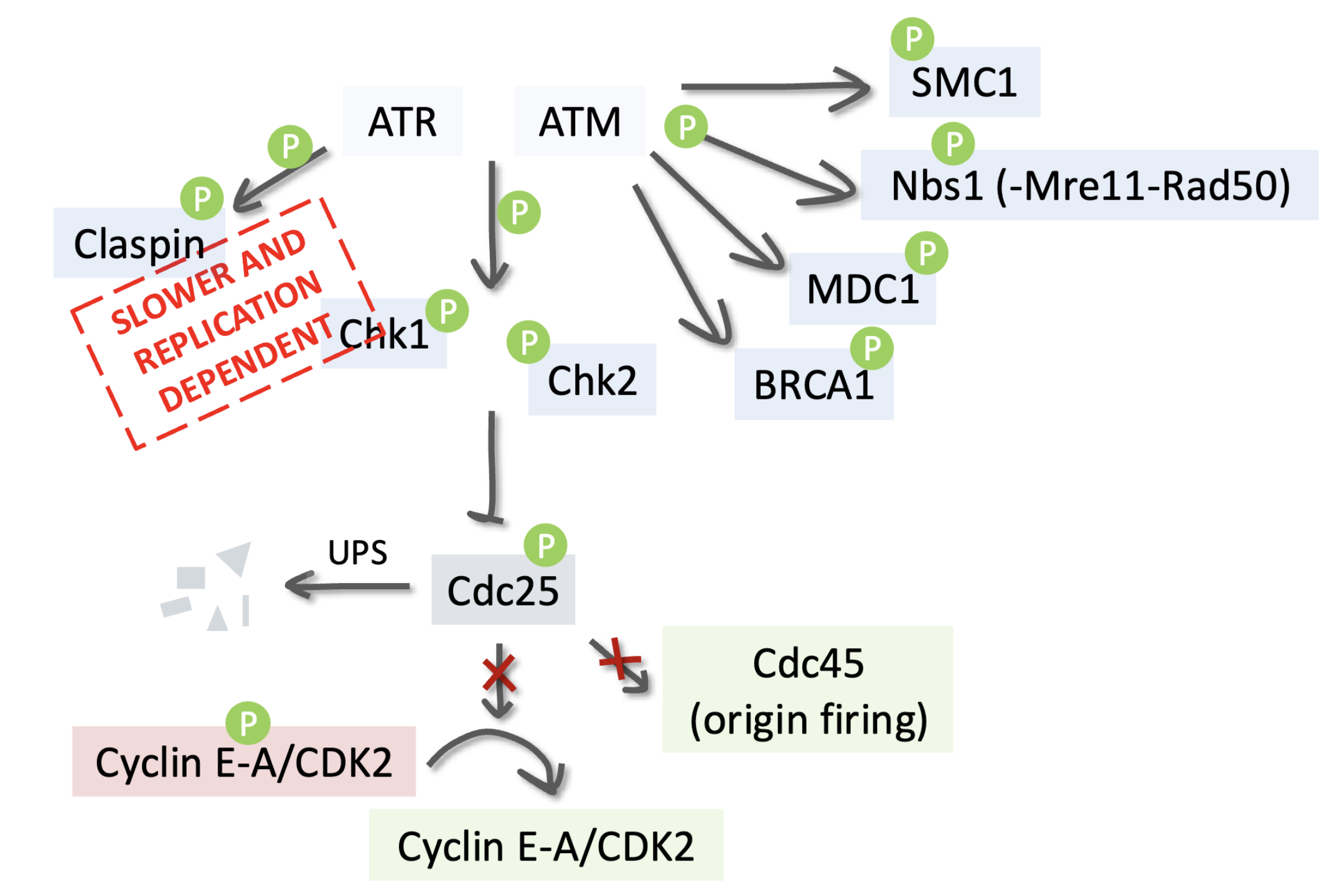
ATR is triggered by replication stress, ssDNA, and stalled forks (replication-dependent)
Claspin, a mediator protein, helps ATR activate Chk1 by phosphorylation
Activated Chk1 phosphorylates Cdc25 phosphatase
Phosphorylated Cdc25 is targeted for ubiquitin-mediated degradation (UPS)
Without Cdc25, Cyclin E/A–CDK2 complexes remain inactive
CDK2 activity drops → new origin firing (via Cdc45) is prevented → DNA replication pauses
Stalled forks are stabilised, no new replication origins fire, and the cell buys time to repair or restart replication.
ATM is activated by MRN complex at DSBs and has a faster response (replication independent) and it phosphorylates many key DDR proteins:
SMC1 – involved in sister chromatid cohesion
Nbs1 (part of MRN complex) – reinforces ATM signaling
MDC1 – recruits more repair factors to damage sites
BRCA1 – coordinates DNA repair via homologous recombination
Chk2 – another checkpoint kinase (parallel to Chk1)
Both Chk1 (via ATR) and Chk2 (via ATM) phosphorylate and inhibit Cdc25, inhibiting Cyclin E/A–CDK2 activity to block DNA synthesis/S-phase progression, also inhibiting origin firing (Cdc45) which prevents new replication forks from starting while damage is unresolved
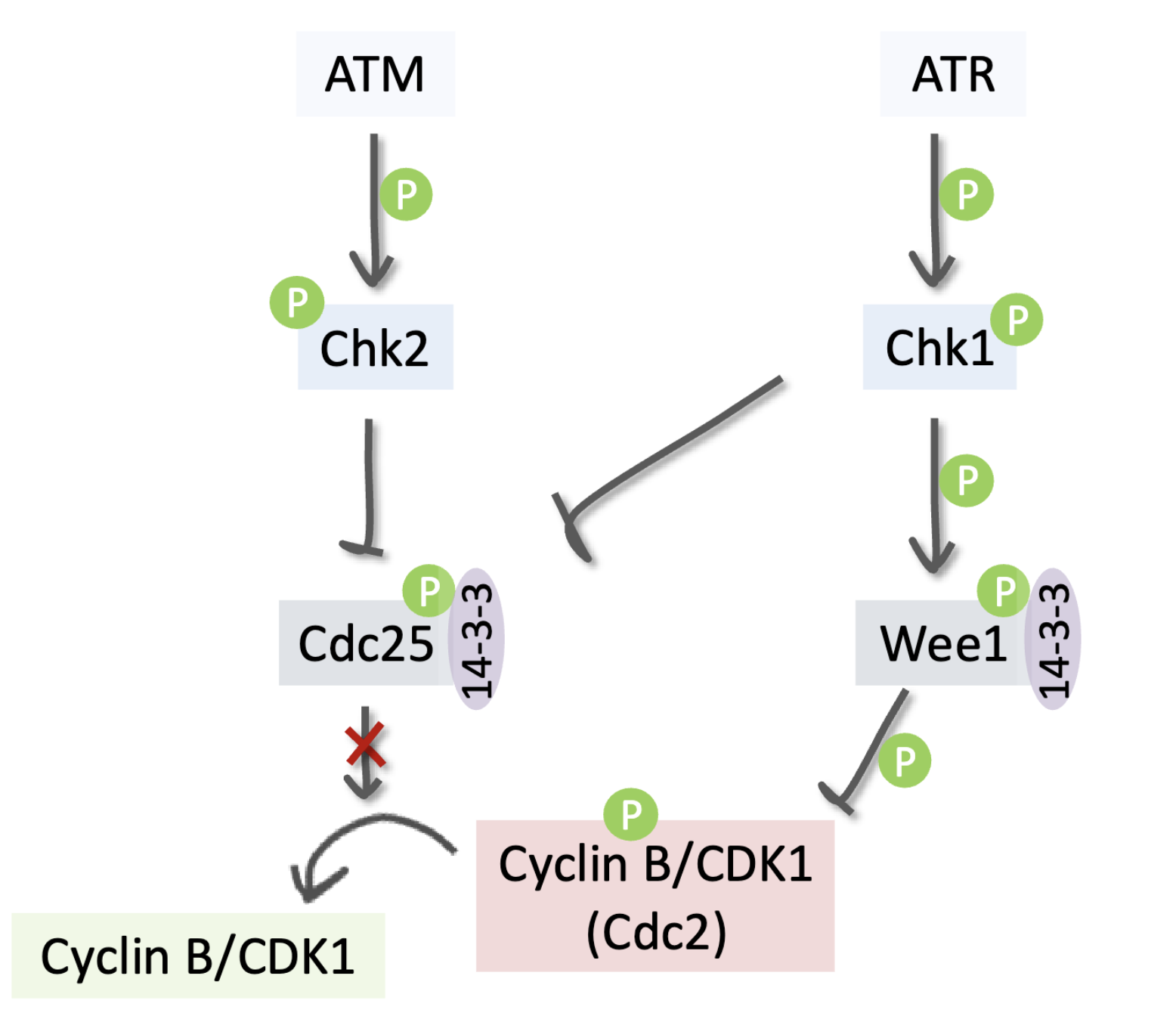
G2/M checkpoint - G2 arrest
Cancer cells strongly depend on this to prevent death by mitotic catastrophe
ATM phosphorylates/activates Chk2 which inhibits Cdc25, preventing it from activating Cyclin B/Cdk1
ATR phosphorylates/activates Chk1 which phosphorylates/ activates Wee1, allowing it to inhibit Cyclin B/Cdk1 via inhibitory phosphorylation
Spindle assembly checkpoint (mitotic checkpoint phase)
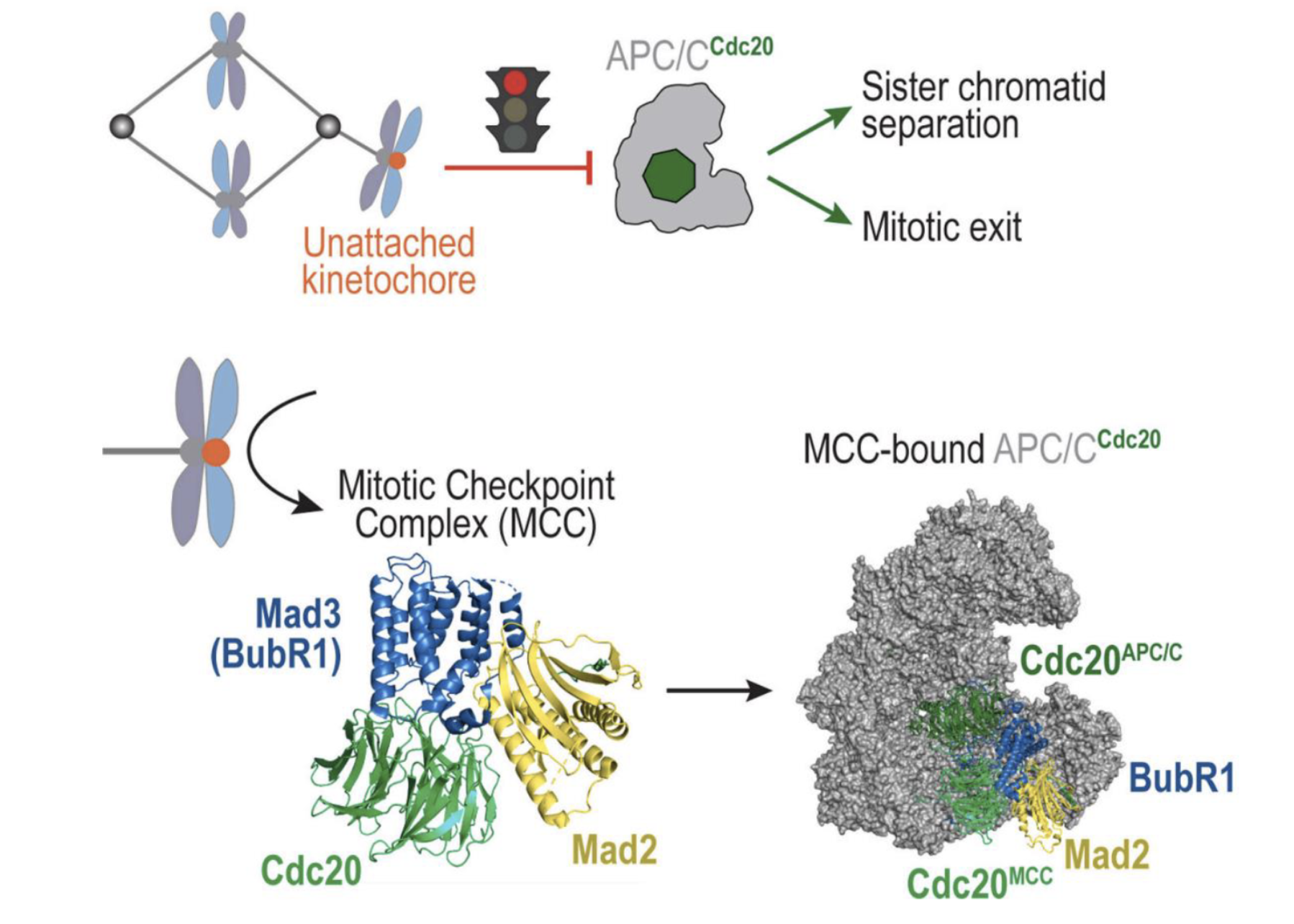
APC/C (Anaphase-Promoting Complex/Cyclosome) is an E3 ubiquitin ligase that drives the cell from metaphase to anaphase.
Its co-activator, Cdc20, helps it ubiquitinate key substrates: securin (inhibitor of separase), cyclin B
When APC/C–Cdc20 is active securin is degraded → separase cleaves cohesin → sister chromatids separate
Cyclin B is also degraded → mitotic exit occurs
When a kinetochore is unattached it recruits checkpoint proteins which form the mitotic checkpoint complex (MCC) - BubR1, Bub3, Cdc20, and Mad2
MCC binds and inhibits APC/C-Cdc20 and prevents it from targeting securin and cyclin B for degradation
Cell cycle checkpoint dysregulation in cancer
This can be leveraged to induce synthetic lethality
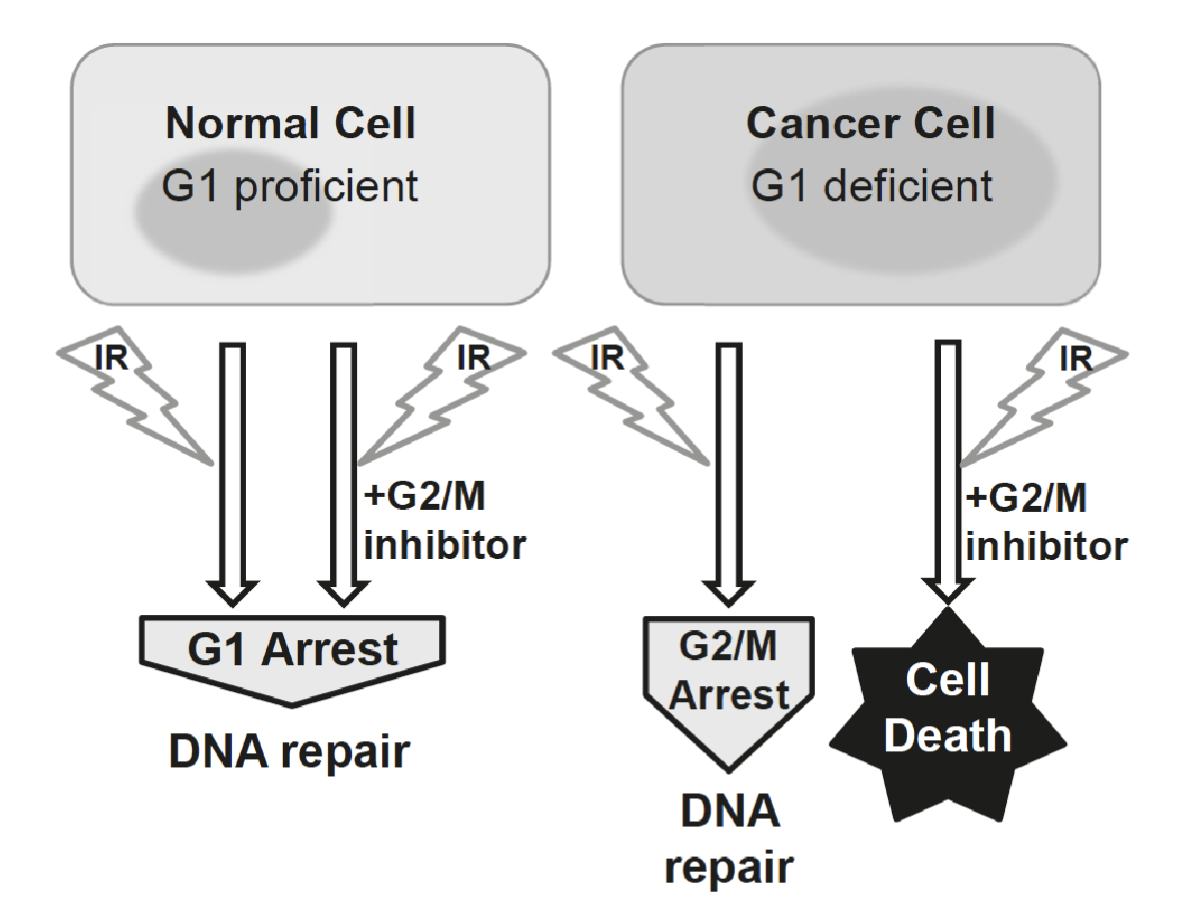
In normal cells:
IR causes DNA double-strand breaks (DSBs).
The G₁ checkpoint, governed mainly by p53 → p21 → CDK inhibition, arrests the cell cycle before S phase.
This allows DNA repair mechanisms (like NHEJ and HR) to fix the damage.
The cell survives and resumes the cycle once repaired
In cancer:
Many cancer cells have defective G₁ checkpoints because they have lost p53 function.
They cannot pause at G₁ after DNA damage.
Instead, they rely heavily on the G₂/M checkpoint, controlled by Chk1, Chk2, and Cdc25 regulation, to pause before mitosis and repair DNA damage
IR still induces a G₂ arrest for DNA repair, however If you treat G₁-deficient cancer cells with ionizing radiation (IR) and a G₂/M checkpoint inhibitor (e.g., an ATR, Chk1, or Wee1 inhibitor), the G₂/M checkpoint is disabled, the cell cannot repair DNA before mitosis, causing mitotic catastrophe and cell death
Studying cell cycle
Yeast genetics
Basic organisation of the cell cycle is essentially the same in all eukaryotes
Many important discoveries about cell cycle control have come from systematic searches for mutations in yeast that inactivate gene encoding essential components of cell cycle control system
Cell free models
Gentle centrifugation can break open large batch of frog eggs and separate cytoplasm from other cell components
Undiluted cytoplasm is collected and sperm nuclei are added with ATP
Sperm nuclei decondense and undergo DNA replication and mitosis, showing that cell cycle control system is operating in the cell free cytoplasmic extract
Mammalian cell models
Divisions are slower than cell free models
Measuring cell cycle progression
Micropscopy - cell shape, staining with DNA binding fluorescent dyes or antibodies, incorporation of nucleotide analogues
Measure of DNA content - flow cytometry
FACS analysis when staining for DNA
Propidium iodide (PI) intercalates into DNA and fluoresces proportionally to the amount of DNA in each cell
Cells can be pulsed with BrdU which is a synthetic analog of thymidine and an anti-BrdU antibody with fluorescent tag could be used to measure DNA replication
During mitosis, Histone H3 becomes phosphorylated on serine 10 (Ser10), so an antibody specific for phospho-H3 (Ser10) can be used to label mitotic cells
Fluorescent, ubiquitination based cell cycle indicator (FUCCI)
FUCCI uses two fluorescently tagged cell cycle–regulated proteins that are degraded at specific phases of the cell cycle, CDT1 (licensing factor that marks G₁ phase, degraded at onset of S pahse) and geminin (inhibitor of CDT1, present during S, G₂, and M phases, degraded at the end of mitosis)
hCdt1(1/100): red fluorescence
hGeminin(1/110): green
G1: red (CDT1 only), G2: yellow (CDT1 ↓, Geminin ↑), G2/M: green (geminin only)
Replicative DNA synthesis (RDS) assay - measures how much DNA replication continues after DNA damage, especially after exposure to ionizing radiation (IR) or UV light
Radio-resistant DNA synthesis
[3H]thymidine incorporation into DNA decreases inc ells that have fucntional checkpoint response
High [3H]thymidine incorporation indicates deficient intra-S checkpoint
DNA fibre assay
CldU (Chloro-deoxyuridine) → labeled with red fluorescence (e.g., anti-BrdU antibody that recognizes CldU)
IdU (Iodo-deoxyuridine) → labeled with green fluorescence (different antibody that distinguishes it from CldU)
Cells are first incubated with CldU (red) for a defined time (e.g., 20 min).
Then switched to IdU (green) for another period (e.g., 20 min).
DNA fibers are spread on glass slides (called “DNA combing”), fixed, and stained with antibodies that recognize each label.
Under a fluorescence microscope, you see colored tracks corresponding to replicated DNA.
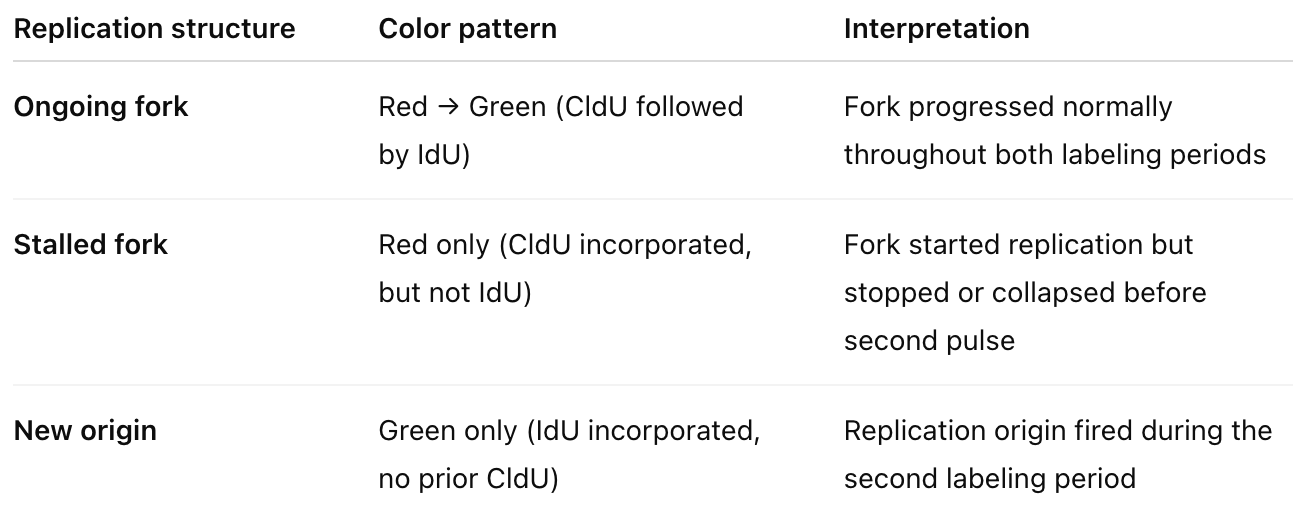
From this we can determine fork speed, stalling frequency, new origin firing rate, fork restart efficiency, asymmetry between sister forks