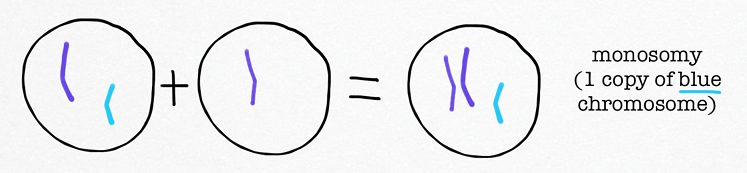Honors Bio Semester 2
Honors Biology Final Exam Review Page
Unit: Energy in Cells
Topic 1: Metabolism- Chapter 6 pages 122-140
Describe the role of exergonic and endergonic reactions in metabolism.
Exergonic - take out energy
Endergonic - create / take in energy
Describe how cells store and release energy through the ATP/ADP cycle.
Release energy through breaking bonds
Store energy by creating them
Topic 2: Cellular Respiration- Chapter 7 pages 141-160
Describe the structure of the mitochondria and how it facilitates aerobic cellular respiration.
Folds - lots of surface area for cellular respiration to take place
Describe the transfer of electrons during redox reactions and be able to identify in a reaction pair which is oxidized and which is reduced.
Redox Reactions: Involve the transfer of electrons between molecules.
Oxidation: Loss of electrons.
Oxidized molecule: Loses electrons.
Reduction: Gain of electrons.
Reduced molecule: Gains electrons.
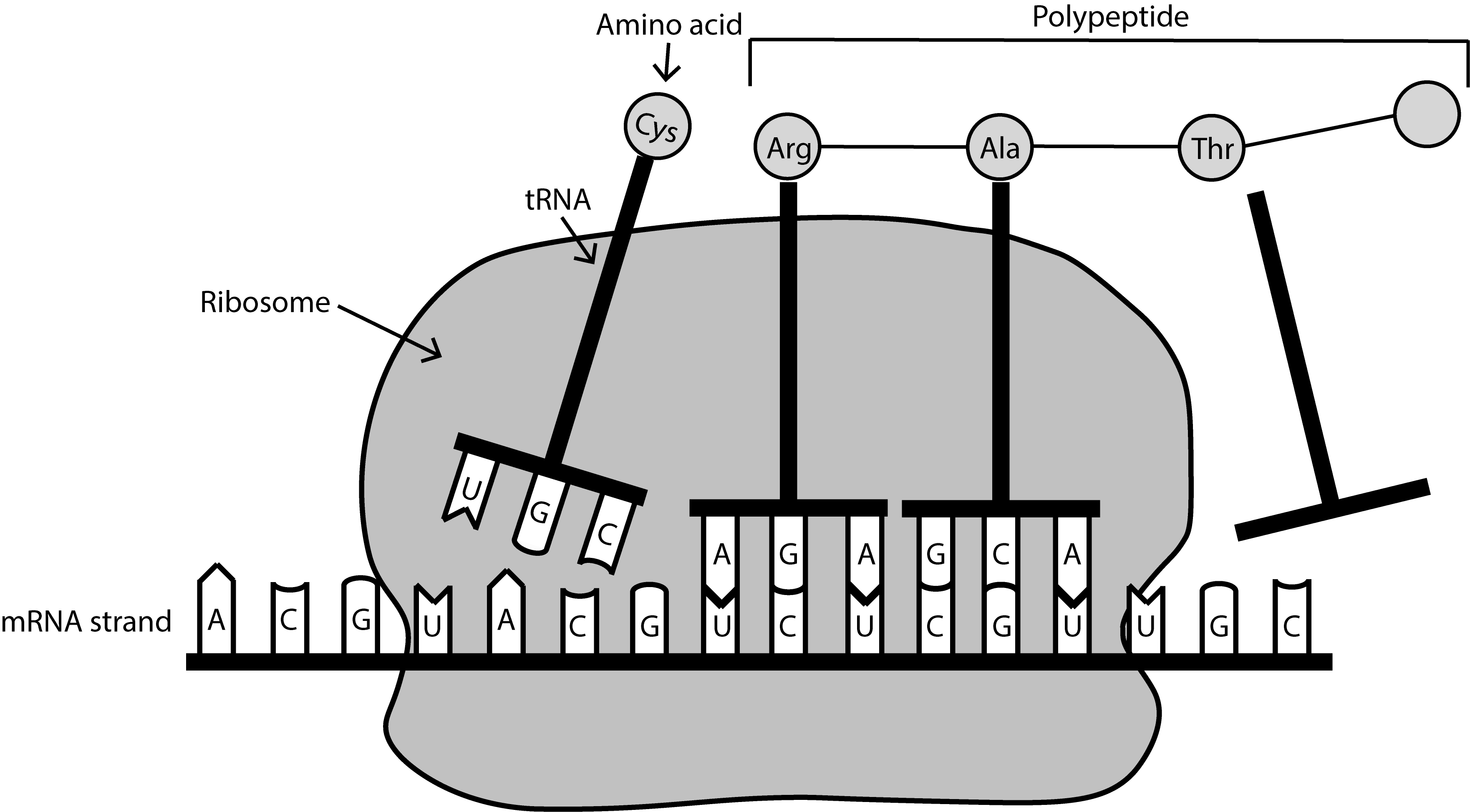
Describe how electron carriers facilitate the transfer of energy during cellular respiration.
Electron Carriers: Molecules that transport electrons during cellular respiration.
Examples: NAD++ (Nicotinamide adenine dinucleotide), FAD (Flavin adenine dinucleotide) Role: They pick up electrons during oxidation reactions.
Transfer electrons to the Electron Transport Chain (ETC) where ATP is produced.
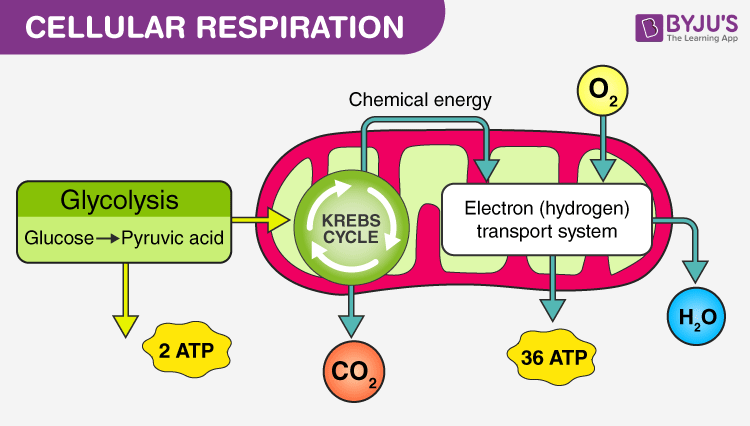
Describe the three stages of Cellular Respiration- Glycolysis, Krebs Cycle and the Electron transport chain and identify the following at each stage:
Glycolysis
Location: Cytoplasm.
Reactants: Glucose, 2 NAD+, 2 ATP.
Products: 2 Pyruvate, 2 NADH, 4 ATP (net gain of 2 ATP).
Electron Carriers: NADH.
2. Krebs Cycle (Citric Acid Cycle)
Location: Mitochondrial matrix.
Reactants: Acetyl-CoA, NAD+, FAD.
Products: 2 CO2, 3 NADH, 1 FADH2, 1 ATP per Acetyl-CoA (per cycle; 2 cycles per glucose molecule).
Net Gain: 2 ATP (from two cycles per glucose molecule).
Electron Carriers: NADH, FADH2.
3. Electron Transport Chain (ETC)
Location: Inner mitochondrial membrane.
Reactants: NADH, FADH2, O2.
Products: H2O, ATP.
Net Gain: About 34 ATP.
Electron Carriers: NADH, FADH2.
Compare alcohol and lactic acid fermentation with aerobic respiration.
Alcohol Fermentation
Occurs In: Yeasts and some bacteria.
Process:
Glycolysis produces 2 ATP.
Pyruvate is converted to ethanol and CO2.
NADH is oxidized back to NAD+ to be reused in glycolysis.
Products: Ethanol, CO2.
Lactic Acid Fermentation
Occurs In: Muscle cells, some bacteria.
Process:
Glycolysis produces 2 ATP.
Pyruvate is converted to lactic acid.
NADH is oxidized back to NAD+ to be reused in glycolysis.
Products: Lactic acid.
Aerobic Respiration
Occurs In: Most eukaryotic cells.
Process:
Glycolysis produces 2 ATP.
Pyruvate enters mitochondria, converted to Acetyl-CoA.
Krebs cycle and ETC produce additional ATP.
Total ATP yield: About 38 ATP per glucose molecule.
Products: CO2, H2O, ATP.
Differences
Oxygen Requirement: Aerobic respiration requires oxygen; fermentation does not.
ATP Yield: Aerobic respiration yields much more ATP (about 38) compared to fermentation (2 ATP).
End Products: Aerobic respiration produces CO2 and H2O; fermentation produces ethanol and CO2 (alcohol fermentation) or lactic acid (lactic acid fermentation).
Topic 3: Photosynthesis- Chapter 8 pages 161-181
Describe leaf structures critical to photosynthesis and explain the role of each. 🍃
Chloroplasts: Contain chlorophyll, the pigment that captures light energy.
Stomata: Pores on the leaf surface for gas exchange, allowing CO2 to enter and O2 to exit.
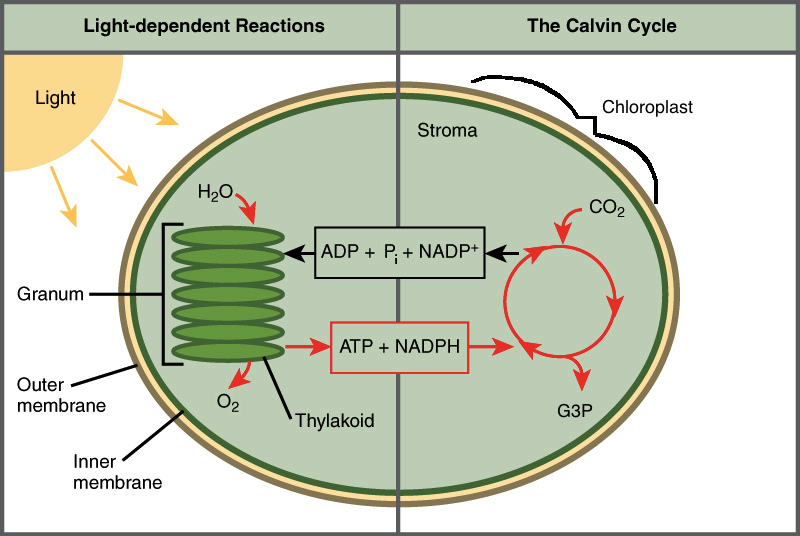
Describe the structure of the chloroplast and how it captures and converts light energy into stored chemical energy through photosynthesis.
Thylakoid Membranes: Contain chlorophyll and other pigments, where light reactions occur.
PANCAKES! 🥞
More pancakes = more spots to catch light = more production
Stroma: Fluid-filled space surrounding thylakoids, where the Calvin cycle takes place.
SYRUP SPACE! 🚀
Chlorophyll: Absorbs light energy and initiates the process of photosynthesis.
Describe and model the two stages of photosynthesis (the light reactions and the Calvin cycle) including where each occurs and the products and reactants of each.
Light Reactions: 🌕
Occur in thylakoids.
Reactants: Light, water, ADP, NADP+.
Products: ATP, NADPH, oxygen.
Calvin Cycle (Dark Reactions): 🌑
Occur in the stroma.
Reactants: ATP, NADPH, CO2.
Products: Glucose (sugar), ADP, NADP+
Describe factors that impact the rate of photosynthesis.
Light Intensity: More light increases the rate of photosynthesis until it plateaus
Carbon Dioxide Concentration: Higher CO2 levels increase photosynthesis until it plateaus.
Temperature: Optimal temperatures enhance enzyme activity; extremes inhibit photosynthesis.
Describe the relationship between photosynthesis and cellular respiration.
Photosynthesis converts light energy into chemical energy (glucose) and releases oxygen.
Cellular respiration breaks down glucose to release energy for cellular processes and produces CO2 and water as byproducts.
They are complementary processes: the products of one are the reactants of the other.
Unit: DNA Structure and Replication, Cell Cycle
Topic 1: DNA; Chapter 13, pages 253-277
Targets: I can…
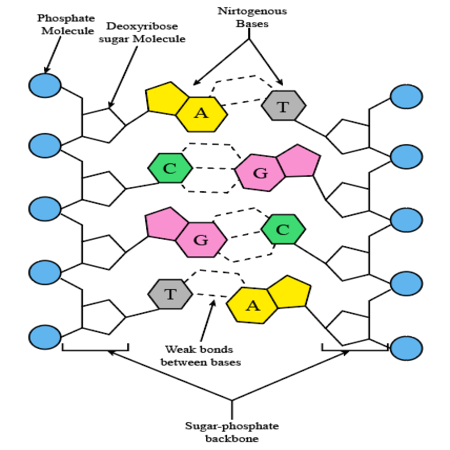
Describe the composition and structure of a nucleic acid using DNA and RNA as models.
Both DNA and RNA are polymers made up of nucleotides.
Each nucleotide consists of a sugar molecule (deoxyribose in DNA, ribose in RNA), a phosphate group, and a nitrogenous base (adenine, guanine, cytosine, and thymine in DNA; adenine, guanine, cytosine, and uracil in RNA).
Nucleotides are linked together through phosphodiester bonds to form long chains.
Describe differences between RNA and DNA molecules.
RNA is single-stranded, while DNA is double-stranded.
RNA contains ribose sugar, while DNA contains deoxyribose sugar.
RNA contains uracil as a nitrogenous base instead of thymine found in DNA
Describe how and why DNA bases pair complementary.
Adenine pairs with thymine (A-T), forming two hydrogen bonds.
Guanine pairs with cytosine (G-C), forming three hydrogen bonds.
Complementary base pairing ensures the accurate replication of DNA during cell division.
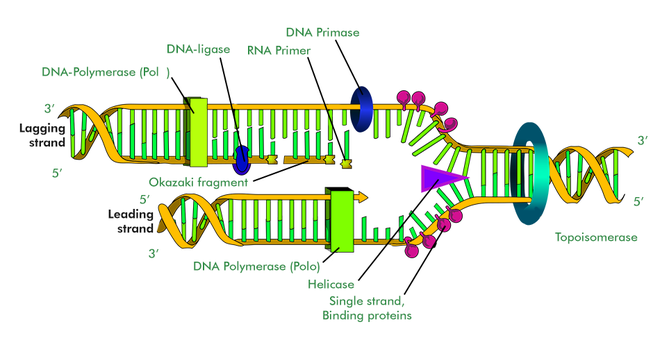
Describe and model the steps of semiconservative replication of DNA.
Initiation: DNA helicase unwinds the double helix, creating replication forks.
Elongation: DNA polymerase adds complementary nucleotides to the template strands, synthesizing new DNA strands in the 5' to 3' direction.
Termination: DNA polymerase continues until it reaches the end of the DNA molecule or encounters another replication fork.
Describe the 5’ and 3’ ends of a DNA molecule and how this determines its structure.
The 5' end has a phosphate group attached to the 5' carbon of the sugar molecule.
The 3' end has a hydroxyl (-OH) group attached to the 3' carbon of the sugar molecule.
The directionality of DNA is determined by the orientation of the sugar-phosphate backbone, with one end having a free phosphate group (5') and the other end having a free hydroxyl group (3').
Describe and model continuous replication of the leading strand and discontinuous replication of the lagging strand of DNA.
The leading strand is synthesized continuously in the 5' to 3' direction toward the replication fork.
The lagging strand is synthesized discontinuously in short fragments called Okazaki fragments, which are later joined by DNA ligase.
Identify and describe the roles of the following enzymes that facilitate DNA replication- Helicase, Primase, DNA polymerase, Ligase.
Helicase: Unwinds the DNA double helix.
Primase: Synthesizes RNA primers on the template DNA strands.
DNA Polymerase: Adds complementary nucleotides to the growing DNA strand.
Ligase: Joins Okazaki fragments on the lagging strand and seals nicks in the DNA backbone.
Describe how the parental strand and complementary strand of a DNA molecule are related.
The parental DNA strand serves as a template for the synthesis of a new complementary DNA strand.
The complementary DNA strand is built based on the sequence of nucleotides in the parental DNA strand, following the rules of complementary base pairing.
NOT IDENTICAL TO PARENT STRAND YOU KNOW BETTER ⏰🆘 👿👹👺🔥
Topic 2: The Cell Cycle and Mitosis; Chapter 9, pages 182-198, Chapter 16, pages 321-341
HS-LS1-4. Use a model to illustrate the role of cellular division (mitosis) and differentiation in producing and maintaining complex organisms.
Targets: I can…
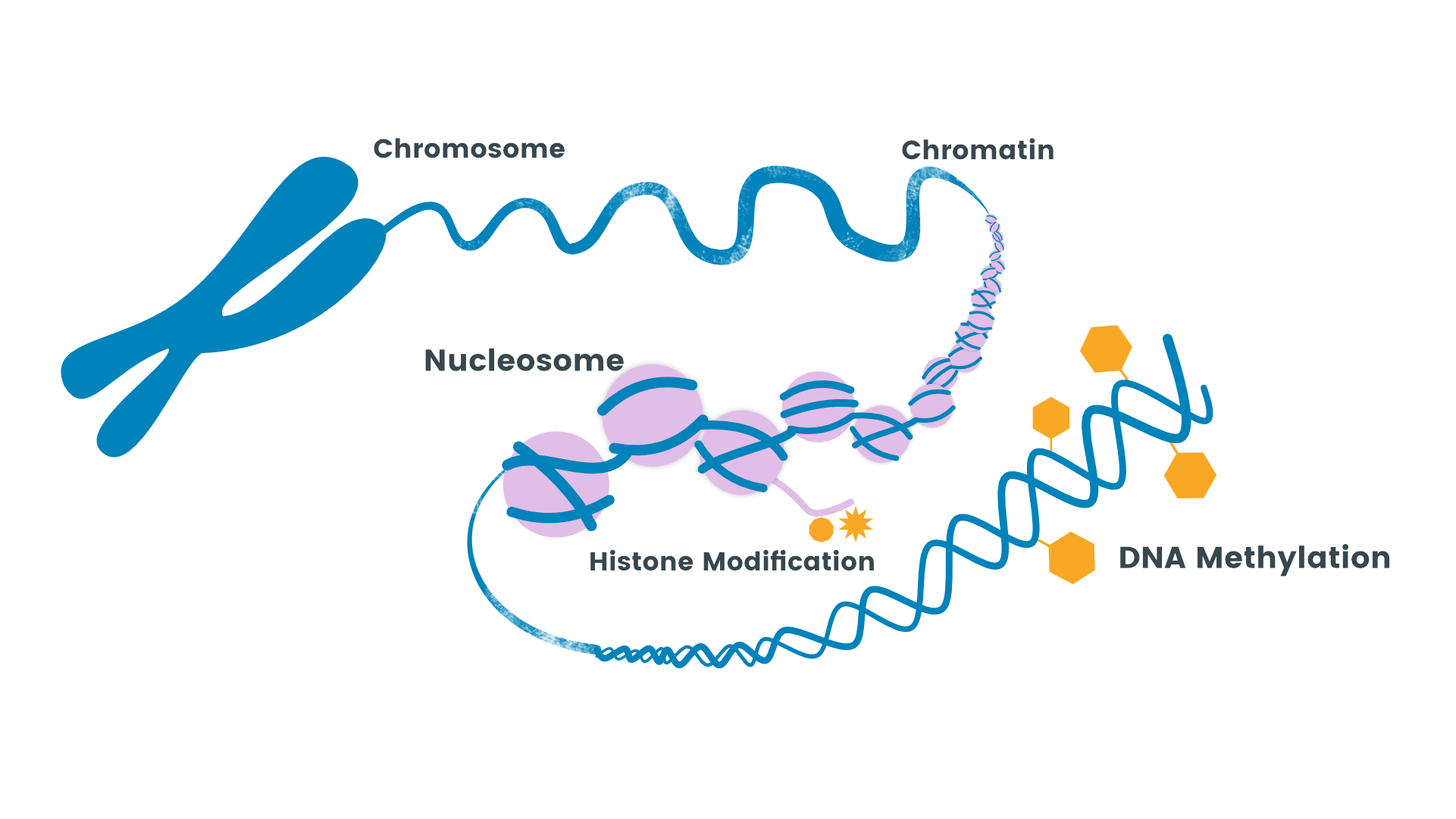
Describe and model the cellular organization of genetic material using the terms - genome, gene, chromosomes, sister chromatids and chromatin.
Genome: Complete set of an organism's DNA
Gene: Segment of DNA that codes for a specific protein or RNA molecule.
Chromosomes: Thread-like structures made of DNA and proteins (histones) that carry genetic information.
Sister Chromatids: Identical copies of a chromosome, formed during DNA replication and joined at the centromere.
Chromatin: Complex of DNA and proteins (histones) that condenses to form chromosomes during cell division.
Explain the importance of the surface area to volume ratio in the division of cells.
A high surface area to volume ratio allows for efficient exchange of materials (such as nutri.ents and waste) with the environment.
As cells grow larger, their volume increases faster than their surface area, leading to decreased efficiency in material exchange.
Cell division (such as mitosis) maintains an optimal surface area to volume ratio by producing smaller daughter cells with higher surface area to volume ratios.
Describe the major occurrences in the GO, G1, S and G2 phases of interphase.
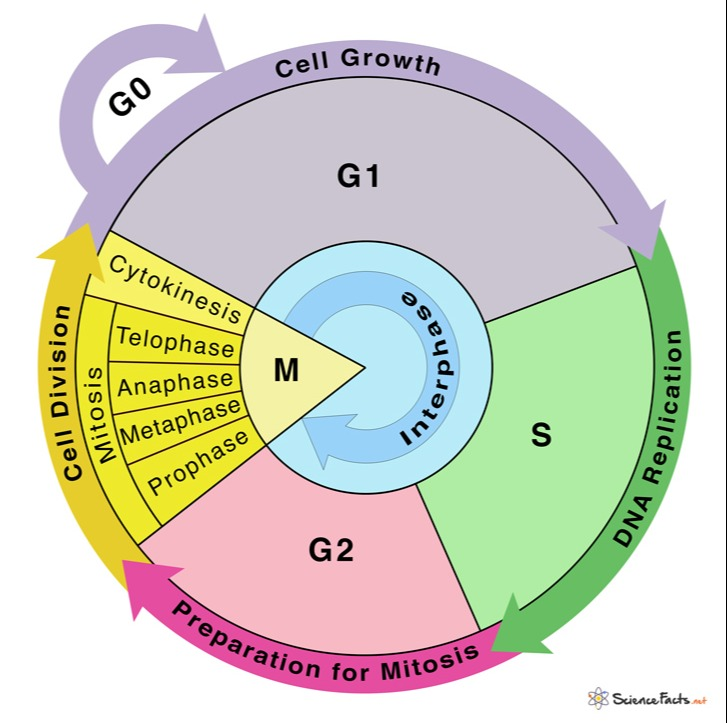
G0 Phase: Non-dividing phase where cells exit the cell cycle and may enter a resting state.
G1 Phase: Cell grows and carries out normal functions; prepares for DNA replication.
S Phase: DNA replication occurs, resulting in the synthesis of sister chromatids.
G2 Phase: Cell continues to grow and prepares for mitosis; checks for DNA errors and repairs any damage.
Describe and model the major occurrences in each phase of mitosis in plant and animal cells.
Prophase: Chromosomes condense, spindle fibers form, nuclear envelope breaks down.
Metaphase: Chromosomes align at the cell's equator (metaphase plate).
Anaphase: Sister chromatids separate and move toward opposite poles of the cell.
Telophase: Chromatids arrive at opposite poles, nuclear envelope reforms, chromosomes decondense.
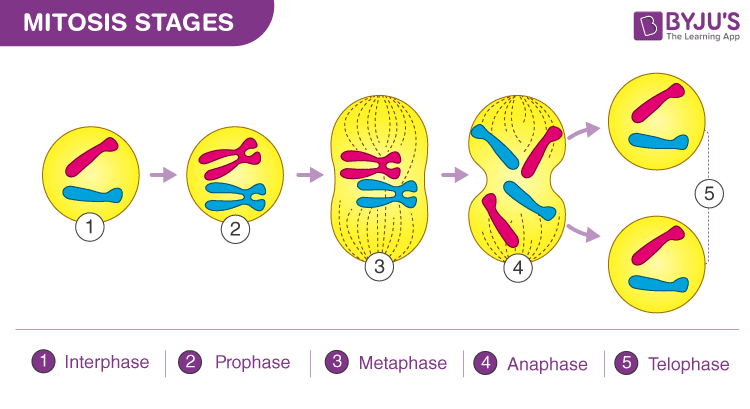
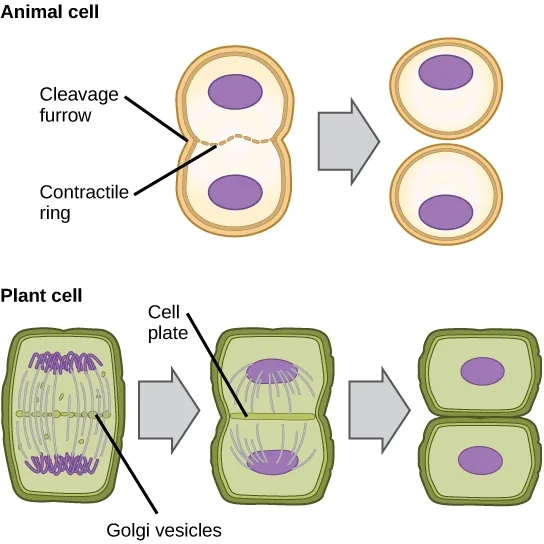
Compare a parent cell with the resulting daughter cells produced from mitosis.
Parent cell is diploid (2n), while daughter cells are genetically identical and also diploid (2n).
Daughter cells have the same number of chromosomes as the parent cell.
Daughter cells may be slightly smaller due to cytokinesis, the division of cytoplasm.
Describe the role of mitosis in producing and maintaining complex organisms.
Mitosis ensures the growth, repair, and maintenance of tissues and organs by producing genetically identical daughter cells.
It allows organisms to develop and replace damaged or old cells, contributing to overall organismal homeostasis and complexity.
Unit: Protein Synthesis, Cancer, Cell Differentiation and Epigenetics
Chapter 6 pages 131-138; Chapter 14 pages 278-300; Chapter 16 pages 321-340
HS-LS1-1. Construct an explanation based on evidence for how the structure of DNA determines the structure of proteins which carry out the essential functions of life through specialized cells.
Targets: I can…
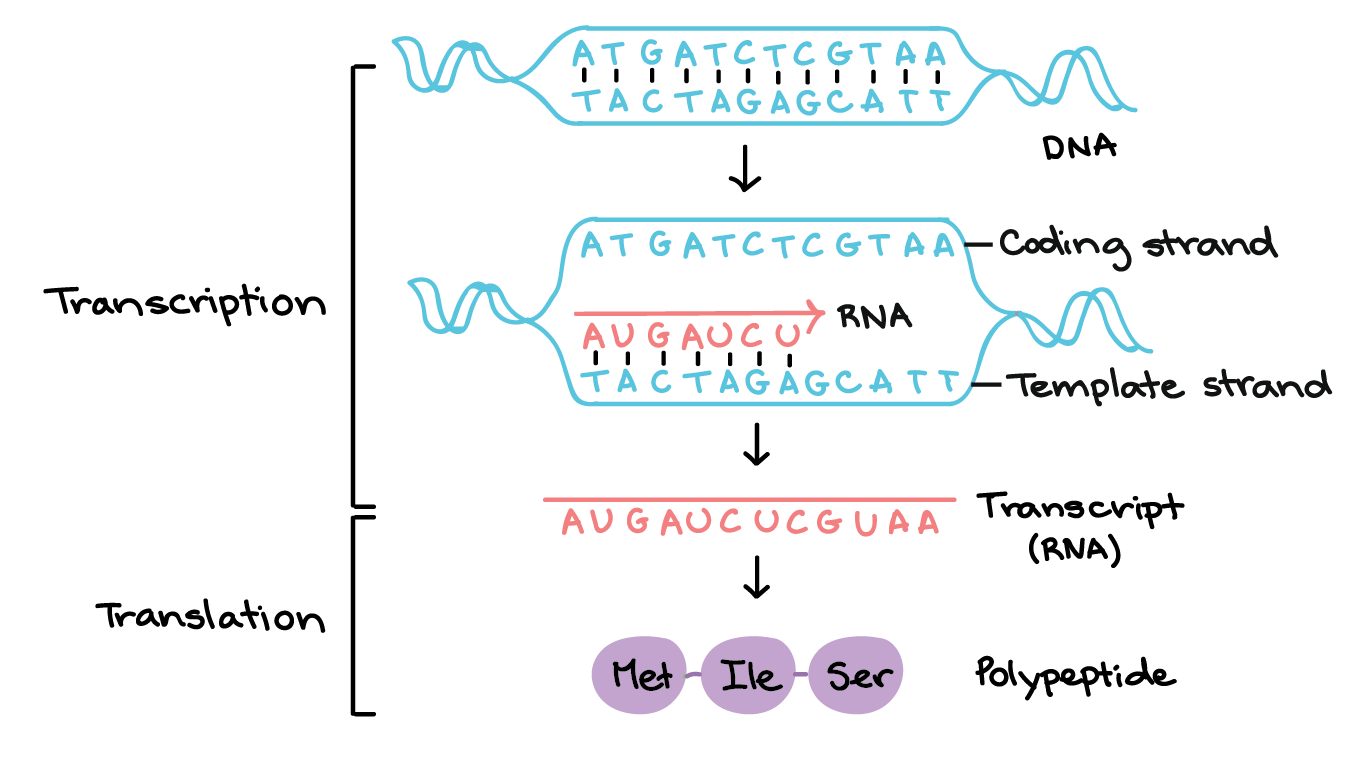
Describe and model the relationship among the following molecules - amino acid, polypeptide, and protein.
Amino acids are the building blocks of proteins.
A polypeptide is a chain of amino acids linked by peptide bonds.
Proteins are complex molecules made up of one or more polypeptide chains folded into specific three-dimensional structures.
Describe the difference between the primary and quaternary structure of proteins, and explain how both relate to the function of a protein.
Primary Structure: Linear sequence of amino acids in a polypeptide chain.
Quaternary Structure: Arrangement of multiple polypeptide chains (subunits) in a protein.
Primary structure determines the sequence of amino acids, while quaternary structure involves interactions between different polypeptide chains.
Both structures are crucial for the function of a protein: primary structure determines the overall shape, while quaternary structure allows for complex functions such as enzyme activity or structural support.
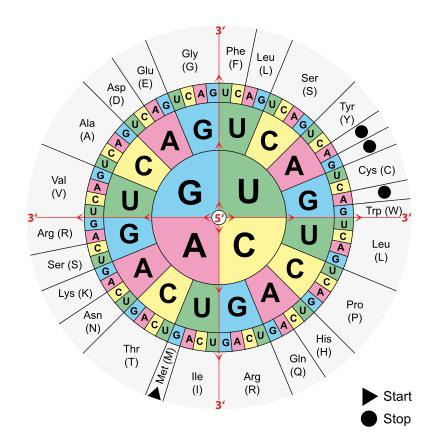
Describe the major occurrences in the three steps of protein synthesis (transcription, RNA processing, and translation) and the role of genes, DNA, mRNA, tRNA, rRNA, codons, anti-codons and ribosomes.
Transcription: DNA sequence is transcribed into mRNA by RNA polymerase.
RNA Processing: Pre-mRNA is edited (e.g., splicing) to produce mature mRNA.
cut out introns, fuse exons (edit - express)
Translation: mRNA is translated into a polypeptide chain by ribosomes with the help of tRNA and rRNA.
Describe how the following mutations can occur and how they would affect resulting gene sequences- substitution (silent, missense, and nonsense) and frameshift (addition, deletion and nucleotide-pair deletion).
Substitution: Replaces one nucleotide with another.
Silent: No change in amino acid sequence.
Missense: Results in a different amino acid.
Nonsense: Creates a premature stop codon, leading to a truncated protein.
Frameshift: Insertion or deletion of nucleotides, causing a shift in the reading frame.
Addition: Inserts one or more nucleotides.
Deletion: Removes one or more nucleotides.
Nucleotide-pair deletion: Removes two adjacent nucleotides.
Mutations can alter the amino acid sequence of a protein, affecting its structure and function.
Describe the role of post-transcriptional control of gene expression in an organism. (alternative splicing)
Alternative splicing allows different mRNA isoforms to be produced from the same gene.
This process can generate multiple protein variants with different functions or regulatory properties from a single gene.
Guac cirlce 😎
Chapter 15 pp 308-309; Utah Genetics webquest.
HS-LS1-1. Construct an explanation based on evidence for how the structure of DNA determines the structure of proteins which carry out the essential functions of life through specialized cells.
Targets: I can...
Describe the role of pre-transcriptional (DNA) control of gene expression in an organism.
Pre-transcriptional control refers to mechanisms that regulate gene expression before the process of transcription begins.
It involves factors that determine whether a gene is accessible for transcription by RNA polymerase or is kept in a repressed state.
Examples include DNA methylation, histone modifications, and the binding of regulatory proteins to specific DNA sequences (such as enhancers and silencers).
Pre-transcriptional control plays a crucial role in determining which genes are expressed and at what levels, thereby influencing cellular functions and responses to environmental stimuli.
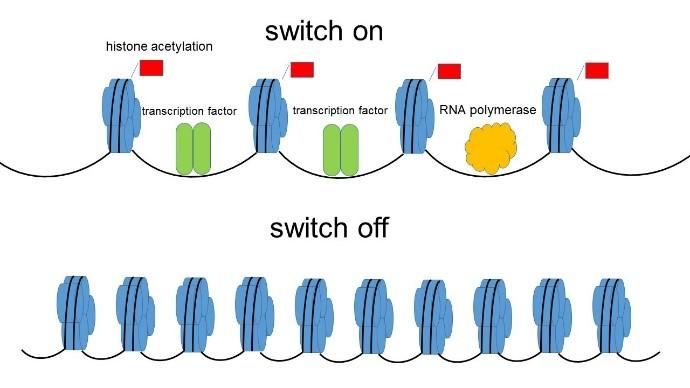
Describe the regulation of chromatin structure and gene expression through histone modifications and DNA methylation and acetylation.
Chromatin structure can be modified through histone modifications, such as methylation, acetylation, phosphorylation, and ubiquitination.
DNA methylation involves the addition of methyl groups to cytosine bases, which can repress gene expression by blocking the binding of transcription factors or recruiting proteins that promote chromatin condensation.
Histone acetylation neutralizes the positive charge of histone proteins, loosening the chromatin structure and allowing access to DNA for transcription factors and RNA polymerase.
These modifications regulate gene expression by controlling the accessibility of DNA sequences to transcriptional machinery, thereby influencing which genes are transcribed and at what levels.
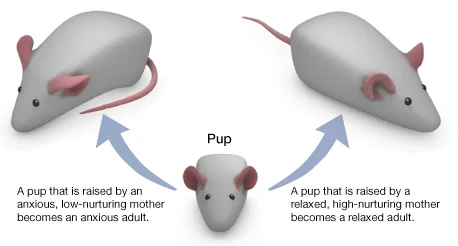
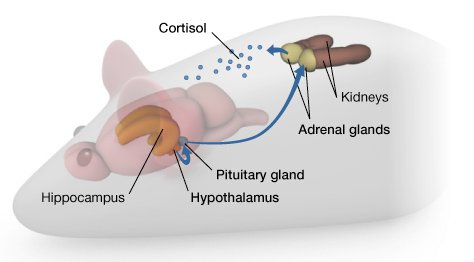
Describe the effects of the “Lick your rats” experiment in regard to gene expression.
Rats that received high levels of maternal care (licking and grooming) exhibited reduced stress response and increased expression of genes associated with stress regulation in the hippocampus compared to rats with low maternal care.
This study highlighted the role of environmental factors, such as maternal care, in shaping gene expression patterns and behavioral outcomes, providing insights into the mechanisms underlying gene-environment interactions.
Describe what the epigenome is and how it is different from the genome.
The epigenome refers to the overall pattern of chemical modifications to DNA and histone proteins that regulate gene expression without altering the underlying DNA sequence.
Unlike the genome, which encompasses the entire genetic material of an organism encoded in its DNA sequence, the epigenome consists of reversible chemical modifications that control when and where genes are expressed.
While the genome remains relatively stable throughout an organism's lifetime, the epigenome is dynamic and can be influenced by various environmental factors, developmental cues, and lifestyle choices.
Analyze the effects of temperature on pigment production in Serratia marcescens.
Serratia marcescens is a bacterium known for producing a red pigment called prodigiosin, which is influenced by environmental conditions such as temperature.
Higher temperatures can stimulate the production of prodigiosin, leading to increased pigment production.
This phenomenon may be attributed to temperature-dependent regulation of gene expression or enzymatic activity involved in prodigiosin biosynthesis pathways.
Understanding the effects of temperature on pigment production in Serratia marcescens can provide insights into the molecular mechanisms underlying bacterial physiology and adaptation to different environmental niches.
Abnormal Cell Division
Chapter 16.3 Pages 334-337
Describe abnormal cellular division that can lead to cancer.
Cancer is characterized by uncontrolled cell growth and division, resulting in the formation of malignant tumors.
Abnormal cellular division in cancer can be caused by mutations or dysregulation of genes involved in cell cycle control, DNA repair, and apoptosis.
Factors such as environmental exposures, genetic predisposition, and viral infections can contribute to the development of cancer.
Abnormalities in key regulatory pathways, such as those involving oncogenes and tumor suppressor genes, can disrupt the normal balance between cell proliferation and cell death, leading to cancerous growth.
Describe the role of oncogenes and tumor suppressor genes in the regulation of the cell cycle.
Oncogenes: These are genes that promote cell growth and division when mutated or overexpressed. They can be proto-oncogenes that regulate normal cell proliferation but become oncogenes when mutated, leading to uncontrolled cell growth.
Examples of oncogenes include growth factor receptors (e.g., EGFR), signal transduction proteins (e.g., RAS), and transcription factors (e.g., MYC).
Oncogenes promote cell cycle progression by activating cell growth pathways, inhibiting apoptosis, or enhancing cell proliferation.
Tumor Suppressor Genes: These are genes that inhibit cell growth and division or promote apoptosis when functioning normally. Loss or inactivation of tumor suppressor genes can contribute to cancer development.
Examples of tumor suppressor genes include p53, RB, and PTEN.
Tumor suppressor genes regulate the cell cycle by inhibiting cell cycle progression, promoting DNA repair, or inducing apoptosis in response to cellular damage or stress.
Mutations or deletions in tumor suppressor genes can lead to loss of their normal function, allowing uncontrolled cell growth and tumor formation.
Unit: Meiosis and Genetics
Topic 1 Meiosis; Chapter Chapter 10 pages 200-213 and Chapter 12 (section 12.4) pages 248-251
Targets: I can…
Describe the diploid chromosome number in human somatic cells and how sex chromosomes determine gender.
Human somatic cells typically contain 46 chromosomes arranged in 23 pairs, making them diploid (2n).
Sex determination is governed by the presence of sex chromosomes.
Females have two X chromosomes (XX), while males have one X and one Y chromosome (XY).
The presence of the Y chromosome determines male gender.
Describe the haploid number of human chromosomes in a gamete and the importance of these cells being haploid.
Human gametes (sperm and egg cells) contain 23 chromosomes each, making them haploid (n).
Haploid cells are crucial for sexual reproduction as they ensure that the resulting zygote will have the correct diploid chromosome number.
During fertilization, the haploid sperm cell fuses with the haploid egg cell to form a diploid zygote with 46 chromosomes.
Describe the difference between asexual and sexual reproduction and their effect on genetic diversity.
Asexual reproduction involves the production of offspring from a single parent without the involvement of gametes. It results in genetically identical offspring, leading to low genetic diversity.
Sexual reproduction involves the fusion of gametes from two parents to produce genetically diverse offspring with unique combinations of genes from both parents. This promotes genetic diversity and increases the chances of survival in changing environments.
Describe and provide examples of homologous chromosomes, autosomes and sex chromosomes.
Homologous Chromosomes: Paired chromosomes that carry genes for the same traits at corresponding loci. One homologous chromosome is inherited from each parent.
Autosomes: Chromosomes that are not involved in sex determination. In humans, autosomes include chromosomes 1 to 22.
Sex Chromosomes: Chromosomes that determine an individual's sex. In humans, sex chromosomes include X and Y chromosomes.
Describe the human life cycle using the following terms- meiosis, fertilization, egg cell, sperm cell, germ line cell and zygote indicating when cells change from haploid to diploid and vice versa.
Germ Line Cells: Diploid cells in the reproductive organs (ovaries in females, testes in males) undergo meiosis to produce haploid gametes (egg and sperm cells).
Fertilization: Haploid egg and sperm cells fuse to form a diploid zygote.
Zygote: The diploid zygote undergoes mitotic cell divisions to develop into a multicellular organism.
The cycle repeats when germ line cells undergo meiosis to produce haploid gametes.
Describe the major events of Meiosis I and Meiosis II correctly using the following terms- homologous chromosomes, daughter cells, diploid, haploid, and sister chromatids.
Meiosis I: Homologous chromosomes separate, resulting in two daughter cells with half the number of chromosomes (haploid).
Meiosis II: Sister chromatids separate, producing four daughter cells, each with a haploid set of chromosomes.
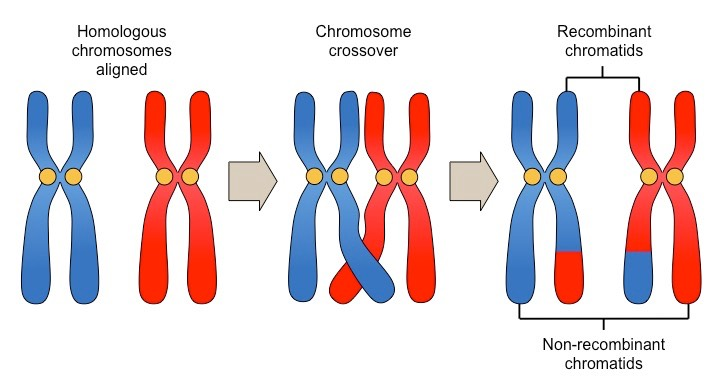
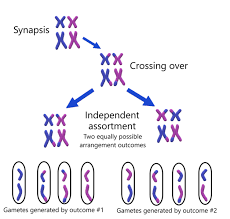
Describe crossing over and independent assortment and their effect on genetic diversity.
Crossing Over: Exchange of genetic material between homologous chromosomes during prophase I of meiosis. This creates new combinations of alleles and increases genetic diversity.
Independent Assortment: Random orientation of homologous chromosomes at the metaphase plate during meiosis I. This leads to the independent inheritance of different genes and further increases genetic diversity.
Describe similarities and differences between mitosis and meiosis.
Similarities: Both processes involve cell division and include stages such as prophase, metaphase, anaphase, and telophase.
Differences: Mitosis produces two identical daughter cells with the same chromosome number as the parent cell (diploid), while meiosis produces four genetically diverse daughter cells with half the chromosome number (haploid).
Describe how and when nondisjunction occurs and how it causes aneuploidy.
Nondisjunction: Failure of homologous chromosomes or sister chromatids to separate properly during meiosis.
Aneuploidy: An abnormal number of chromosomes resulting from nondisjunction. For example, trisomy (three copies of a chromosome) or monosomy (one copy of a chromosome).
Describe and provide causes for monosomic and trisomic zygotes.
Monosomic zygotes result from the fertilization of a gamete lacking a particular chromosome with a normal gamete.
Trisomic zygotes occur when a gamete with an extra chromosome fuses with a normal gamete during fertilization.
Describe the differences between polyploidy and aneuploidy and genetic disorders caused by these conditions.
Polyploidy: A condition in which an organism has more than two complete sets of chromosomes. It can be tolerated in some plant species but usually results in developmental abnormalities and sterility in animals.
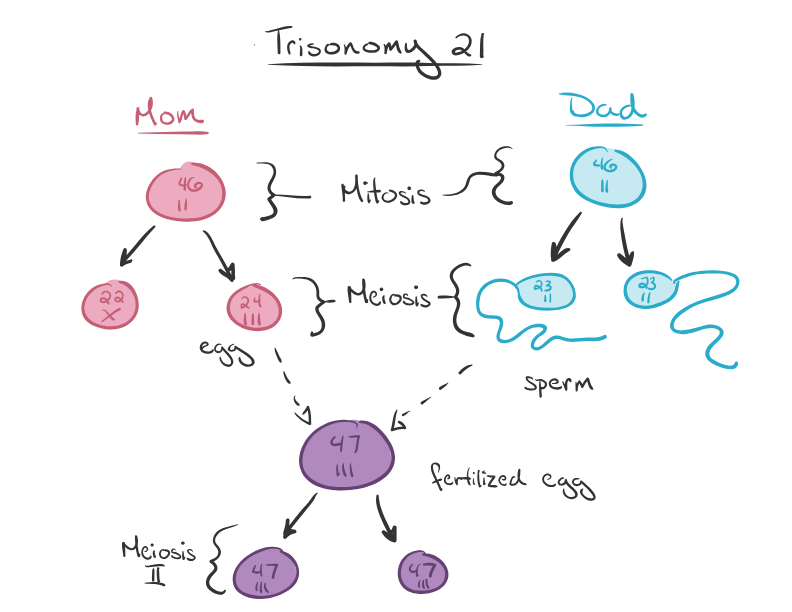
Aneuploidy: A condition in which an organism has an abnormal number of chromosomes, either too many or too few. Aneuploidy can lead to genetic disorders such as Down syndrome (trisomy 21) or Turner syndrome (monosomy X).
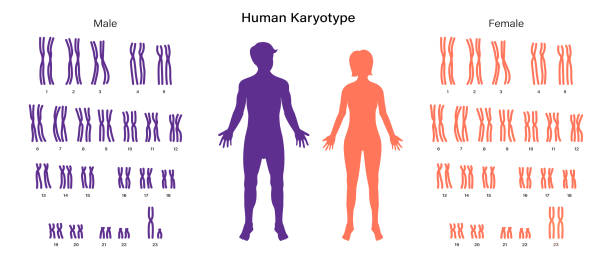
Describe how to construct and interpret a karyotype to determine a patient’s sex, chromosome number, and identify a genetic disorder.
A karyotype is an organized profile of an individual's chromosomes.
It can be used to determine an individual's sex, chromosome number, and identify genetic disorders by examining the size, shape, and banding patterns of chromosomes.
trisomy 21 (Down syndrome) can be identified by the presence of an extra copy of chromosome 21.
Describe deletions, inversions, translocations, and duplications and genetic disorders that may result.
Deletions: Loss of a segment of a chromosome.
Inversions: Reversal of the orientation of a segment within a chromosome. Inversions may disrupt gene function but can also be benign.
Translocations: Movement of a segment from one chromosome to another.
Duplications: Presence of extra copies of a chromosome segment.
Topic 2: Inheritance; Chapter 11 pages 214-235, Chapter 12 pages 236-242
Targets: I can…
Distinguish between an allele and a gene and provide an example of each.
Gene: A unit of heredity that is responsible for determining a particular trait. Genes are located on chromosomes and consist of DNA sequences.
The gene for eye color in humans.
Allele: Different forms of a gene that occupy the same locus on homologous chromosomes and can produce variations in a specific trait.
The gene for eye color may have alleles for blue, brown, or green eyes.
I can describe genotypes and phenotypes using the following terms- dominant, recessive, homozygous, heterozygous, and carrier.
Genotype: The genetic makeup of an organism, represented by the combination of alleles present for a particular trait.
Homozygous dominant (BB), heterozygous (Bb), or homozygous recessive (bb) for the trait of brown eye color.
Phenotype: The observable characteristics or traits of an organism, resulting from the interaction between its genotype and the environment.
Having brown eyes as a result of the genotype BB or Bb.
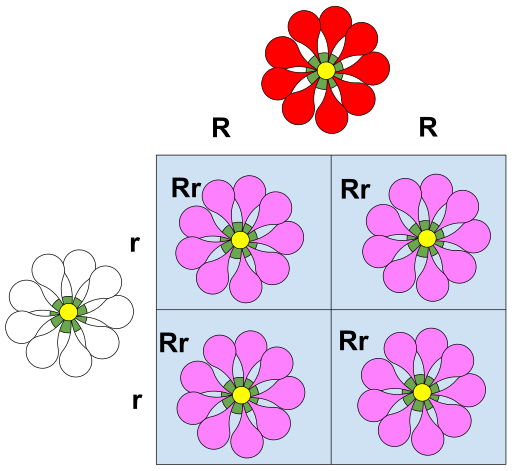
Use punnett squares to determine the probability of phenotypic and genotypic traits in offspring for monohybrid crosses.
Punnett squares are tools used to predict the possible genotypic and phenotypic outcomes of offspring from a cross between two individuals.
For a monohybrid cross involving one trait controlled by a single gene with two alleles (e.g., dominant and recessive), the Punnett square can determine the probability of different genotypes and phenotypes in the offspring.
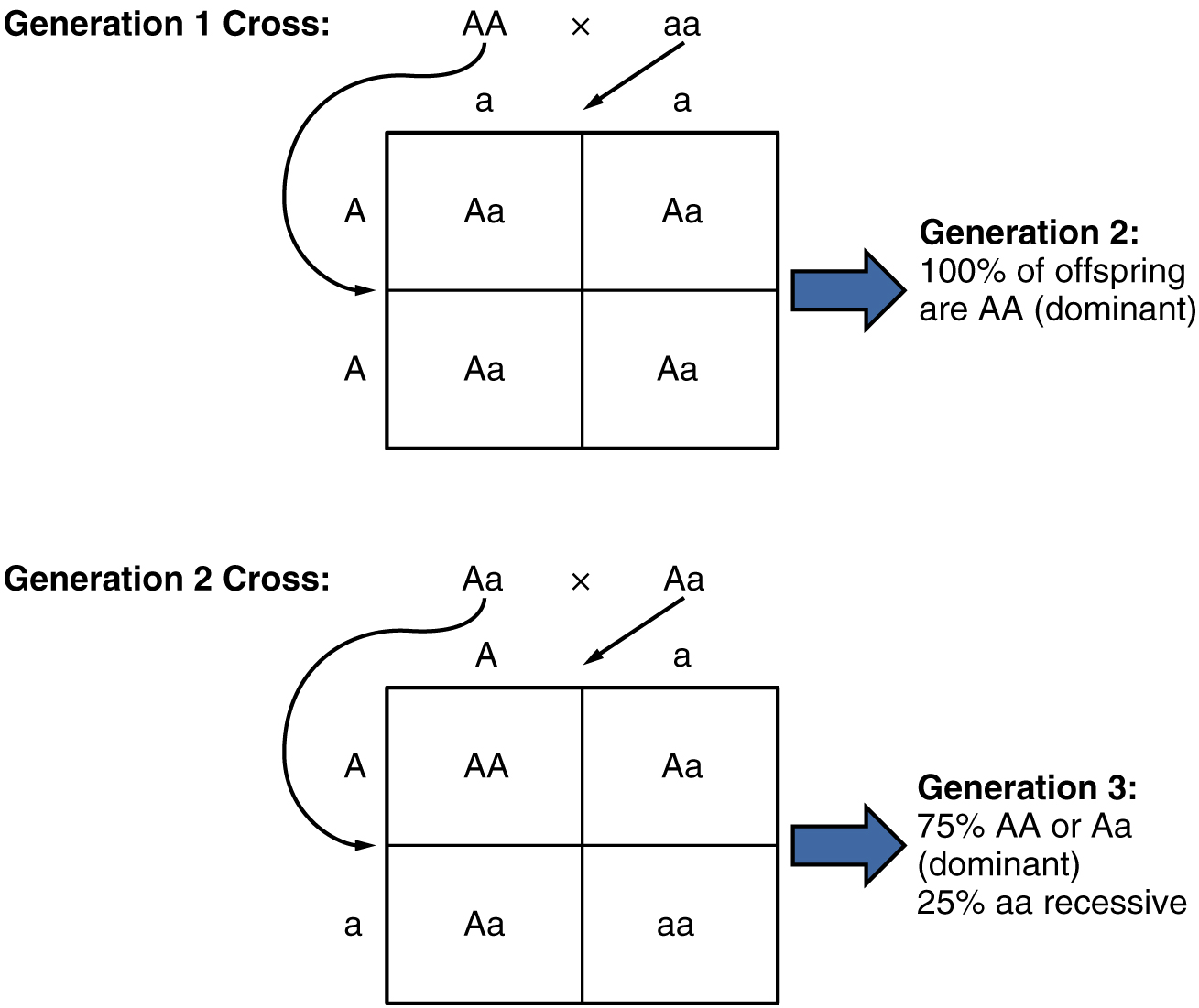
Describe how the laws of probability apply to Mendelian inheritance.
The laws of probability, such as the multiplication and addition rules, apply to Mendelian inheritance to predict the likelihood of specific genotypes and phenotypes in offspring.
The principle of segregation states that alleles segregate randomly during gamete formation, with each gamete having an equal probability of receiving either allele.
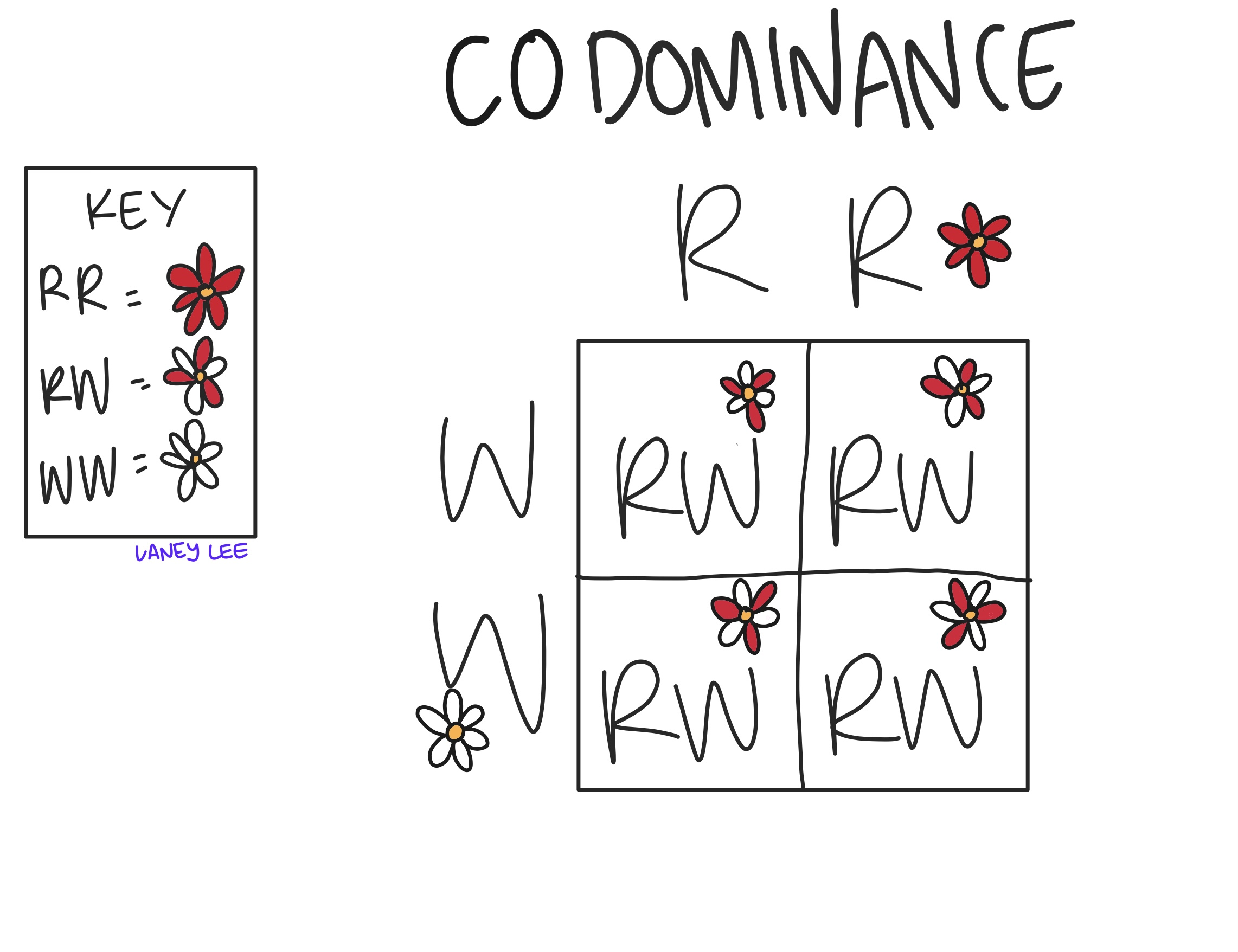
Describe the inheritance of non-mendelian traits including codominance, incomplete dominance, multiple alleles, pleiotropy, polygenics, and epistasis.
Codominance: Both alleles are fully expressed in the heterozygous condition, resulting in a phenotype that shows the traits of both alleles
speckled chicken
Incomplete Dominance: The heterozygous phenotype is an intermediate blend of the two homozygous phenotypes.
grey chicken
Multiple Alleles: When a gene has more than two alleles, each individual still inherits only two alleles, but there are more than two possible alleles in the population.
Epistasis: One gene masks or modifies the effect of another gene at a different locus.
Describe the effect of the environment on the expression of traits.
Environmental factors such as diet, temperature, and exposure to toxins can influence the expression of traits by affecting gene regulation and protein function.
nutrition can impact height or skin color, while temperature can affect fur color in animals.
Describe the inheritance of sex-linked traits.
Sex-linked traits are controlled by genes located on the sex chromosomes (X or Y).
sex-linked traits include color blindness and hemophilia, which are more common in males due to their hemizygous nature (having only one X chromosome).
Read and analyze a pedigree to determine the inheritance pattern of a trait.
Pedigrees are diagrams that show the inheritance pattern of a particular trait or condition within a family over multiple generations.
By examining the pedigree, one can determine the mode of inheritance (autosomal dominant, autosomal recessive, X-linked dominant, X-linked recessive) and predict the likelihood of individuals inheriting the trait.
Anatomy and Physiology
Anatomy and Physiology, Chapters 32.4, 33, and 34 pages 677-730
Explain the pathway of air through the organs in the respiratory system. Describe/identify all organs involved.
Nasal Cavity: Air enters through the nose, where it is filtered, warmed, and humidified.
Larynx: Air moves through the larynx (voice box), which contains the vocal cords.
Trachea: Air travels down the trachea, a tube reinforced with cartilage rings to keep it open.
Bronchi: The trachea splits into two bronchi, one for each lung.
Bronchioles: The bronchi branch into smaller tubes called bronchioles within the lungs.
Alveoli: Air reaches the alveoli, tiny sacs where gas exchange occurs. Oxygen diffuses into the blood, and carbon dioxide diffuses out to be exhaled.
Explain the stages of digestion and the pathway of food through the digestive system. Describe/identify all organs involved.
Mouth: Digestion begins as food is chewed and mixed with saliva, which contains enzymes that start breaking down carbohydrates.
Esophagus: Food moves down the esophagus via peristalsis (wave-like muscle contractions).
Stomach: Food is mixed with gastric juices, breaking down proteins .
Small Intestine:
Duodenum: Chyme mixes with bile (from the liver) and pancreatic juices, which further break down nutrients (smaller chemicals)
Large Intestine: Absorbs remaining water and forms solid waste (feces). Also breaks down into smaller particles
Rectum: Stores feces until they are expelled.
Anus: Feces are expelled from the body.
Explain one example of a negative and positive feedback loop.
Negative Feedback Loop:
Example: Body temperature regulation.
When body temperature rises, the hypothalamus triggers mechanisms like sweating and vasodilation to cool the body down.
When body temperature drops, the hypothalamus triggers shivering and vasoconstriction to warm the body up.
This process maintains homeostasis by counteracting deviations from a set point.
Positive Feedback Loop:
Example: Blood clotting.
When a blood vessel is damaged, platelets adhere to the site and release chemicals that attract more platelets.
This process continues, rapidly forming a clot to seal the wound.
Positive feedback amplifies the response until the clot is formed.
Describe at least one example of how a structure in the body supports its function.
Example: The structure of the alveoli in the lungs.
Structure: Alveoli are tiny, balloon-like sacs with thin walls and a large surface area.
Function: They facilitate efficient gas exchange. The large surface area allows for maximum oxygen absorption into the blood and carbon dioxide removal.
Support: The thin walls of the alveoli allow gases to easily diffuse through them, and their close proximity to capillaries ensures quick and efficient gas exchange.
Unit Evolution
Chapters: 19.1, 19.2, 20.1, 20.2, 20.3, 24.1, 24.2, 24.5 (490-492 only)
Targets: I Can…
Chapter 19 Intro, Sections 1 & 2
Describe the contributions of Charles Darwin to the understanding of evolution.
proposed that species evolve over time through the process of natural selection, where individuals with advantageous traits are more likely to survive and reproduce, leading to changes in the inherited characteristics of populations over generations.
Describe how the process of natural selection leads to the inheritance of desirable traits in a population
Natural selection is the process by which individuals with traits that are better adapted to their environment are more likely to survive and reproduce, passing on those traits to future generations.
In a population, individuals vary in their traits due to genetic variation. Some traits may confer advantages, such as better camouflage or resistance to disease, while others may be disadvantageous.
Individuals with advantageous traits are more likely to survive and reproduce, increasing the frequency of those traits in the population over time.
Through this process, natural selection leads to the inheritance of desirable traits, as individuals with these traits are more successful at passing them on to their offspring, gradually shaping the characteristics of the population.
Explain the relationship between the two drivers of evolution: mutation and natural selection.
Mutation is the ultimate source of genetic variation, introducing new alleles and traits into a population's gene pool.
Natural selection acts on this variation, favoring traits that enhance an organism's fitness in its environment.
Mutations provide the raw material for natural selection to work on, creating genetic diversity within populations.
Natural selection acts as a filter, determining which mutations are advantageous, neutral, or deleterious in a given environment.
Over time, beneficial mutations may increase in frequency through natural selection, leading to evolutionary change and adaptation.
mutation and natural selection are intimately linked in the process of evolution, with mutation providing the variation upon which natural selection acts to drive evolutionary change.
Chapter 20 Sections 1, 2, & 3
Illustrate evolutionary relationships based on the shared characteristics of organisms.
use evidence to interpret and develop phylogenetic trees/cladograms.
Endosymbiosis: 25.1
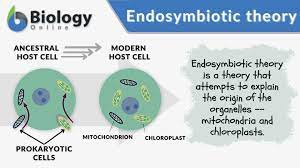
Describe how the theory of endosymbiosis describes important steps between the evolution of prokaryotes to eukaryotes, and explain evidence that supports the theory.
Mitochondria and chloroplasts have their own circular DNA, similar to bacterial DNA.
These organelles replicate independently of the cell through a process resembling binary fission, similar to bacteria.
Mitochondria and chloroplasts have double membranes, consistent with the engulfing mechanism.
Ribosomes in mitochondria and chloroplasts are more similar to bacterial ribosomes than to the eukaryotic cytoplasmic ribosomes.
Microevolution and Macroevolution: 21.1 - 21.4, 22.1 and 22.2, 19.3
Distinguish between microevolution and macroevolution.
Microevolution
Small-scale changes in allele frequencies within a population over a short period.
Example: Changes in the color of a moth population due to pollution.
Macroevolution:
Large-scale evolutionary changes that result in the formation of new species or groups over long periods.
Example: Evolution of mammals from reptilian ancestors
Describe why populations are the smallest biological unit that can evolve.
Evolution involves changes in allele frequencies.
Allele frequencies can only change within populations, not individuals.
Populations contain the genetic variation necessary for natural selection
Describe the concept of evolutionary fitness.
Evolutionary fitness refers to an organism’s ability to survive and reproduce.
High fitness means better adaptation to the environment and more offspring.
Example: A fast predator can catch more prey, increasing its survival and reproduction.
Predict the evolutionary relationship of organisms utilizing anatomical and molecular evidence from both extinct and extant organisms.
Anatomical Evidence: Comparing structures (bones, organs) to determine relatedness.
Molecular Evidence: Analyzing DNA, RNA, or protein sequences to infer evolutionary links.
Both extinct and extant organisms provide data for constructing phylogenetic trees.
Use comparative anatomy and differentiate between homologous, analogous and vestigial structures and embryology.
Homologous Structures: Similar due to shared ancestry.
Example: Forelimbs of humans and bats.
Analogous Structures: Similar due to convergent evolution.
Example: Wings of birds and insects.
Vestigial Structures: Reduced or unused structures from ancestors.
Example: Human appendix.
Evaluate differences in protein sequences (DNA) to create a model that shows the evolutionary relationship between multiple organisms.
Comparing protein sequences (amino acids) helps determine evolutionary relationships.
More similar sequences suggest closer relatedness.
Phylogenetic trees can be built based on these comparisons.
Define and give an example of gene pool.
Gene Pool: Total genetic diversity within a population.
Example: All alleles of the genes in a population of birds.
Describe and explain sources of genetic variation and how they lead to adaptations (morphological, physiological and behavioral).
Mutations: Random changes in DNA.
Gene Flow: Movement of genes between populations.
Genetic Recombination: Shuffling of genes during meiosis.
Variations lead to adaptations:
Morphological: Physical traits (e.g., beak shape).
Physiological: Functional traits (e.g., enzyme efficiency).
Behavioral: Actions or habits (e.g., mating calls).
Compare and contrast natural selection, sexual selection and artificial selection.
Natural Selection: Traits that increase survival and reproduction become more common.
Example: Peppered moth coloration.
Sexual Selection: Traits increase mating success.
Example: Peacock’s tail.
Artificial Selection: Humans select for desirable traits.
Example: Dog breeding.
Compare and contrast stabilizing, directional and disruptive selection.
Stabilizing Selection: Favors intermediate traits.
Example: Human birth weight.
Graph: Narrower peak around the mean.
Directional Selection: Favors one extreme trait.
Example: Giraffe neck length.
Graph: Peak shifts towards one extreme.
Disruptive Selection: Favors both extremes.
Example: Beak sizes in birds.
Graph: Two peaks at both extremes.
Describe a species using the biological species concept and explain how you would determine if two organisms were members of the same species and potential limitations of the concept.
A species is a group of interbreeding organisms that produce fertile offspring.
Determination: Assessing reproductive compatibility.
Limitations: Not applicable to asexual organisms or fossils.
Describe the following isolating mechanisms and explain how they can lead to speciation- geographical, habitat, temporal, behavioral, mechanical, gametic, hybrid viability, hybrid infertility.
Geographical Isolation: Physical separation of populations.
Habitat Isolation: Different habitats within the same area.
Temporal Isolation: Different mating times.
Behavioral Isolation: Different mating behaviors.
Mechanical Isolation: Incompatible reproductive structures.
Gametic Isolation: Sperm and egg cannot fuse.
Hybrid Viability: Hybrids do not survive.
Hybrid Infertility: Hybrids are sterile.
 474 × 262
474 × 262