Prokaryotic Transcription
3/27/24
The Central Dogma
~Genes- the units of heredity- are composed of DNA Sequences required for Gene Expression ~

Genotype: Genes→ RNA→ Phenotype: Proteins
Pyrimidines: simple structure, single ring (Thymine, Cytosine, and Uracil)
Purines: complex structure, double rings (Adenine, and Guanine)
RNA Structure:
Primary Structure
Secondary Stricture: hairpin loop
Tertiary Structure:
What Do All Those RNAs Do?
mRNA-
rRNA-
tRNA
miRNA-
snRNA-
The Molecular Definition of A Gene- all the Dna sequences necessary to synthesize a functional RNA and/or a protein
Genetic Information is NEEDED to:
Recognize a DNA region to be made into an RNA. ARecognice a region withing an mRNA to be translated into a protein
Control the rate og flow of information from DNA→RNA→ Protein (regulatory sequences often termed noncoding, contain “regulatory code” )
Determine the amino acid sequence that codes for a protein (protein code)
The Parts of a Gene:
Prokaryotic genes: Information for multiple proteins is expressed in a single polycistronic mRNA
polycistrinic- an mRNA corresponding to multiple genes whose expression is also controlled by a single promoter and a single terminator
Eukaryotic Genes: A monocistronic mRNA is transcribed from a genes. The code is broken into chunks called exons.
monocistronic- contains only one structural gene, coding for a single polypeptide chain
There is a coding strand running 5’ top 3’
Polymerase reads a 3’ to 5’ template strand to make a 5’ to 3’ RNA
Upstream= 5’ end of stream
Downstream= after the 3’ end of the stream
Rules of Transcription in Prokaryotes
When and Where?: In an undivided compartment, simultaneous with translation
Transcription is a selective process: Only certain parts of DNA are transcribed (downstream of the transcription start )
Promoters: contain short sequences or motifs critical in the binding of RNA polymerase to the DNA strand
Transcription starts at +1 (first nucleotide transcribed)
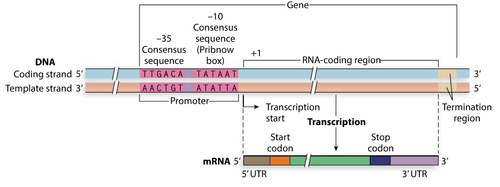
No nucleotide 0
There is no nucleotide 0
+1 is the first nucleotide transcribed
+30 is the thirtieth nucleotide transcribed
-35 is 35 bases before the +1 site
Positions are approximate, there is some variation
Transcription depends on the RNA synthesis by RNA polymerase
RNA polymerase is a multisubunit complex that binds to a promoter and catalyzes the synthesis of RNA from a ssDNA template, no primer needed!
RNA is transcribed form ssDNA
RNA is antiparallel and complimentary to the ssDNA
Transcription is in the 5’ to 3’ direction with respect to the growing molecule adding RNA nucleotide to the 3’ end
First and last bases coded are par of the 5’ UTR and the last bases coded are part of the 3’ UTR
One strand is transcribed per gene, the strand that is the template strand can be different
Which DNA coding strand below would be
associated with the mRNA 5’-CGUACGG-3’?
a) 5’-CGTACGG-3’
b) 5’-GCATGCC-3’
c) 5’-GGCATGC-3’
d) 5’-CCGTACG-3’
Stages of Transcription
Transcription starts with recognition
The promoter is required for transcription to occur.
The RNA Pol Subunits:
core enzyme- polymerase activity
Sigma Subunit- recognizes specific sets of gene
these combine to for the Holoenenzyme- in particular the holoenzyme is attracted to the consensus sequences ( the -35 and the -10)
Initiation is required before Synthesis
Initiation: unwind the DNA to create the transcription bubble; starts with tighter binding forming the closed promoter complex; TATA box is where the DNA will be opened up (the open promoter complex)
Recognition: involves recruitment and binding of the holoenzyme to the promoter and loose binding
Transcription initiation required a template strand (ooen DNA) to make a single stranded RNA
Elongation: Transcription by bacterial RNA polymerase- after the
DNA is unwound ahead of the RNA polymerase by the core enzyme. This maintains the transcription bubble
Termination sequence that come at the end if the 3’ UTR destabilizes the Polymerase
Intrinsic Termination- sequence only
an inverted repeat is a DNA sequence that has two copies each in an opposite direction; inverted repeats cause a 3D structure to form by creating antiparallel and complimentary strands that can hydrogen bond together to form double stranded stem structure, spacer forms a loop - hairpin
Done through the termination sequences
Termination is caused when the hairpin breaks off the rest of the RNA through weak U-T bonds that are weak and allow it to break off.
Rho-Dependent Termination- involves a different termination sequence and a specialized protein that binds to the RNA
Important- Transcription is not a one time event, it will proceed in multiple cycles making more than one copy of the MRNA of a gene. regulation determine how much, when, and where
Imagine you want to prevent transcription of a single bacterial gene by creating a new
mutation in the gene. Which part of the gene would you target for a mutation?
a) Terminator
b) Start codon
c) Promoter
d) Sigma Subunit