BIOCHEMISTRY-ENZYMES & NUCLEIC ACIDS
NOTES FOR SEMIFINALS
Enzyme Basics
Definition: Enzymes are biological catalysts that increase the rate of reactions without being consumed.
Structure: Mostly large, globular proteins with a complex 3D shape.
Catalytic Power: Enzymes lower the activation energy of reactions, allowing them to proceed faster at lower temperatures.
Terms and Definitions
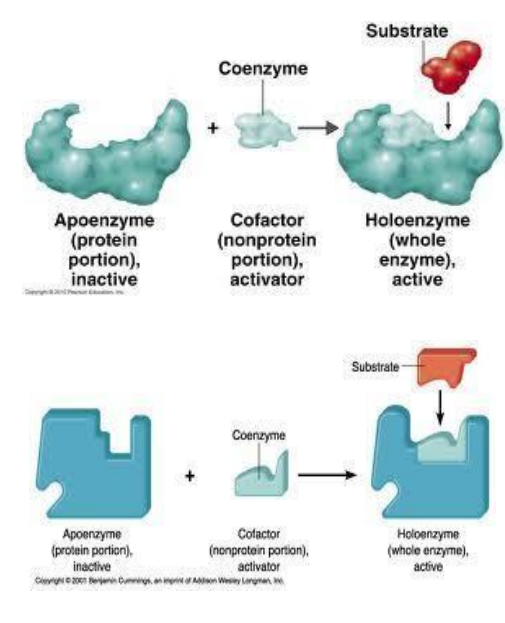
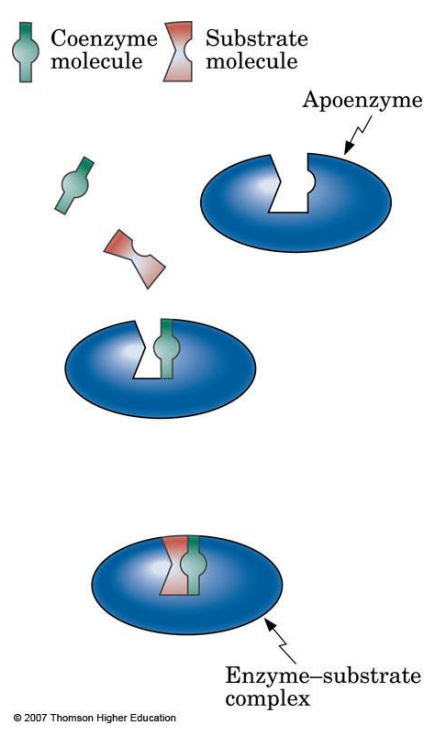
Apoenzyme: Protein part of an enzyme, inactive on its own.
Cofactor: Nonprotein component necessary for enzyme activity; can be metal ions (e.g., Zn²⁺, Mg²⁺).
Coenzyme: Organic cofactor, often a vitamin, assisting in enzyme function.
Substrate: The specific reactant molecule an enzyme acts upon.
Active Site: The specific part of an enzyme where the substrate binds and catalysis occurs.
Catalytic Site: Where the chemical reaction happens.
Binding Site: Holds the substrate in place using weak, non-covalent interactions.
Nomenclature and Classification
Naming: Most enzymes end with "-ase" (e.g., lactase, sucrase).
Some digestive enzymes retain "-in" (e.g., trypsin, pepsin).
Classification: Based on the reaction catalyzed:
Oxidoreductases: Oxidation-reduction reactions.
Transferases: Transfer functional groups.
Hydrolases: Hydrolysis reactions.
Lyases: Add/remove groups without hydrolysis or oxidation.
Isomerases: Rearrange atoms within a molecule.
Ligases: Form bonds with ATP hydrolysis.
Classification of Enzymes
Enzymes are categorized based on the types of reactions they catalyze. Each class has several subclasses that specify the type of reaction even further. The six major enzyme classes are:
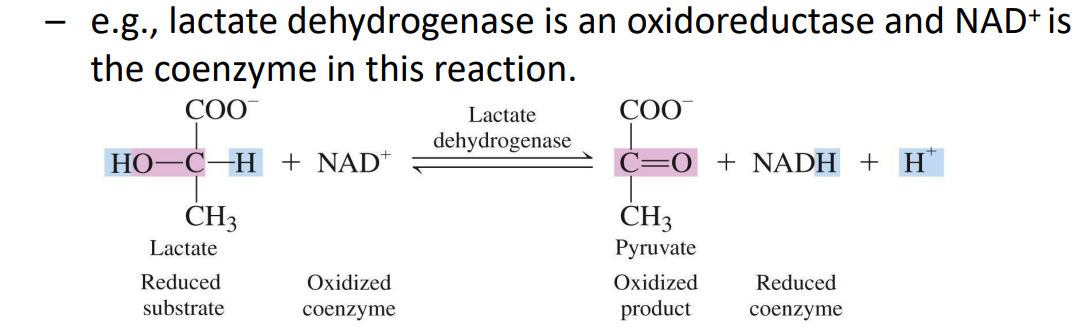
Oxidoreductases – Catalyze oxidation-reduction reactions.
Reaction Type: These enzymes facilitate the transfer of electrons (oxidation-reduction).
Subclasses:
Dehydrogenases: Transfer hydrogen between molecules, often using NAD⁺ or FAD as coenzymes (e.g., lactate dehydrogenase).
Oxidases: Catalyze the transfer of electrons from a substrate to oxygen (e.g., cytochrome oxidase).
Peroxidases: Use hydrogen peroxide as an electron acceptor (e.g., catalase).
Reductases: Catalyze reductions in biochemical pathways.
Hydroxylases: Incorporate hydroxyl groups into substrates by transferring oxygen atoms.
Transferases – Catalyze the transfer of functional groups between molecules.
Reaction Type: Involve the transfer of functional groups, such as methyl, amino, or phosphate groups, from one molecule to another.
Subclasses:
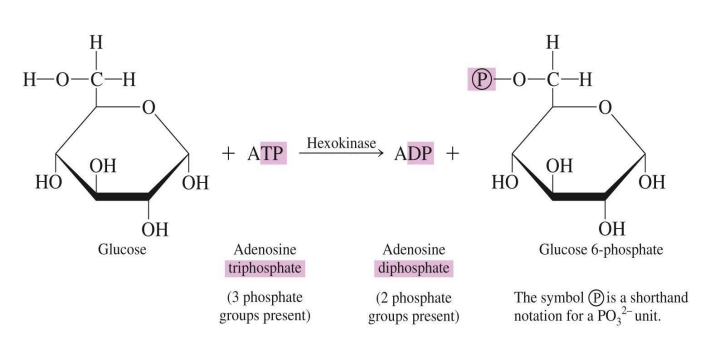
Kinases: Transfer phosphate groups, often from ATP to another molecule (e.g., hexokinase).
Transaminases: Transfer amino groups, especially in amino acid metabolism (e.g., alanine transaminase).
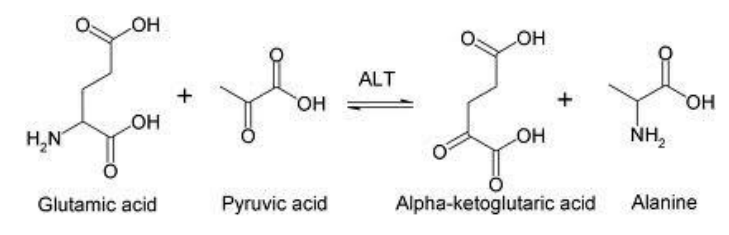
Methyltransferases: Transfer methyl groups (e.g., DNA methyltransferase).
Acyltransferases: Transfer acyl groups (e.g., acetyl-CoA carboxylase).
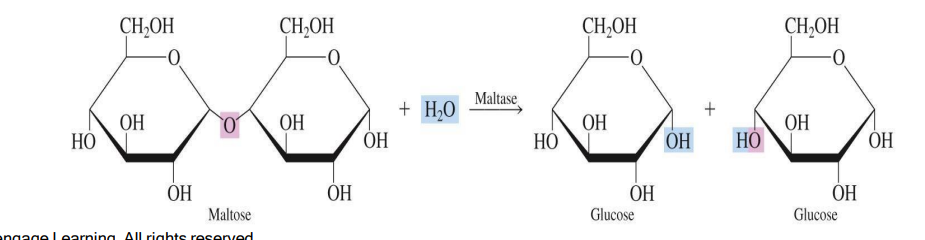
Hydrolases – Catalyze hydrolysis reactions, breaking bonds by adding water.
Reaction Type: Catalyze the cleavage of bonds (C-O, C-N, or C-S) through the addition of water.
Subclasses:
Esterases: Break ester bonds (e.g., lipases, which break down fats).
Proteases (Peptidases): Break peptide bonds in proteins (e.g., trypsin).
Glycosidases: Break glycosidic bonds in carbohydrates (e.g., amylase).
Nucleases: Break phosphodiester bonds in nucleic acids (e.g., DNase).
Phosphatases: Remove phosphate groups from molecules (e.g., alkaline phosphatase).
Lyases – Catalyze the addition or removal of groups to form double bonds or break double bonds without hydrolysis or oxidation.
Reaction Type: Typically add groups to double bonds or remove groups to form double bonds.
Subclasses:
Decarboxylases: Remove carboxyl groups (CO₂) from organic acids (e.g., pyruvate decarboxylase).
Dehydratases: Remove water molecules to form double bonds (e.g., fumarase).
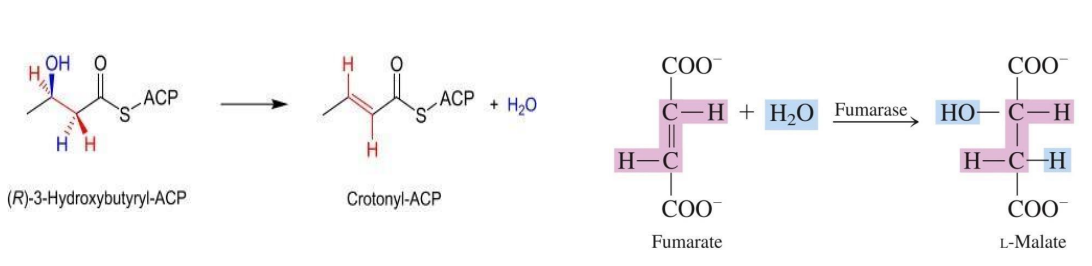
Synthases: Catalyze the joining of two molecules without ATP (e.g., citrate synthase).
Aldolases: Split carbon-carbon bonds (e.g., fructose-bisphosphate aldolase in glycolysis).
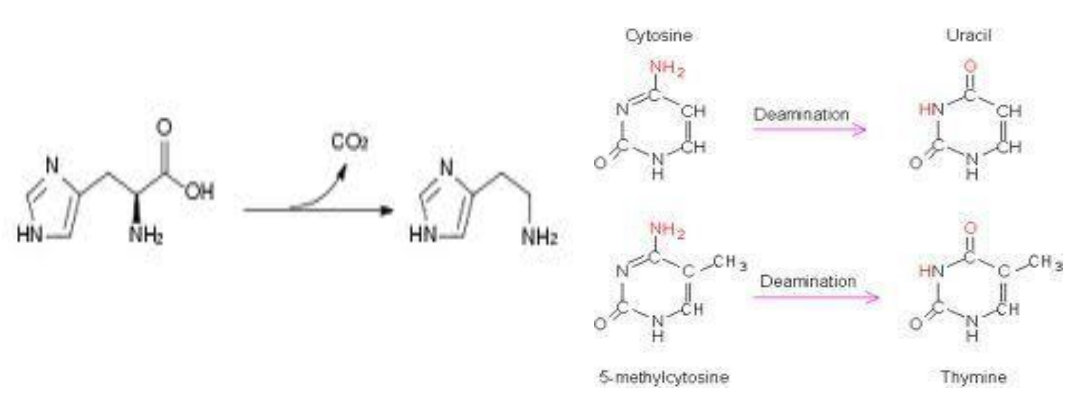
Isomerases – Catalyze isomerization reactions, rearranging atoms within molecules to form isomers.
Reaction Type: Rearrange the molecular structure of substrates to create isomers.
Subclasses:
Racemases and Epimerases: Convert optical or stereoisomers by changing the configuration around a specific carbon (e.g., alanine racemase).
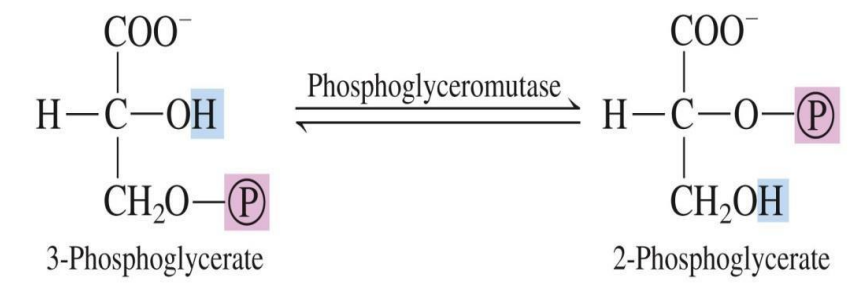
Mutases: Shift functional groups within the same molecule (e.g., phosphoglucomutase).
Cis-trans Isomerases: Convert cis-isomers to trans-isomers or vice versa (e.g., prolyl isomerase).
Ligases – Catalyze the joining of two molecules with covalent bonds, often using ATP.
Reaction Type: Catalyze bond formation between molecules, coupled with ATP hydrolysis.
Subclasses:
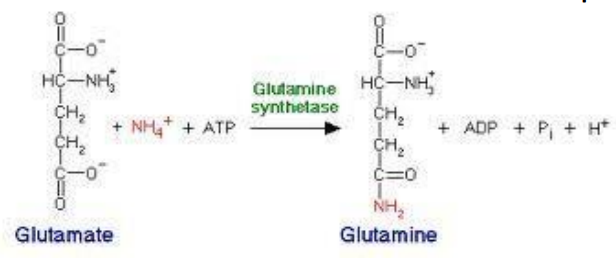
Synthetases: Require ATP to form new bonds between molecules (e.g., glutamine synthetase).
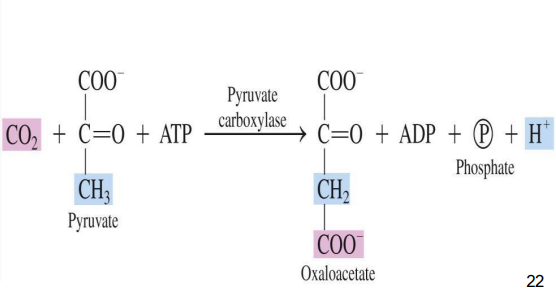
Carboxylases: Add CO₂ to a substrate using ATP (e.g., pyruvate carboxylase).
Ligases: Join two large molecules by forming a new bond, often using ATP (e.g., DNA ligase).
Enzyme Specificity
Absolute Specificity: Acts on a single substrate (e.g., catalase for hydrogen peroxide).
Stereochemical Specificity: Distinguishes between stereoisomers.
Group Specificity: Works on molecules with the same functional group.
Linkage Specificity: Targets specific types of bonds, regardless of molecule structure.
Mechanism of Action
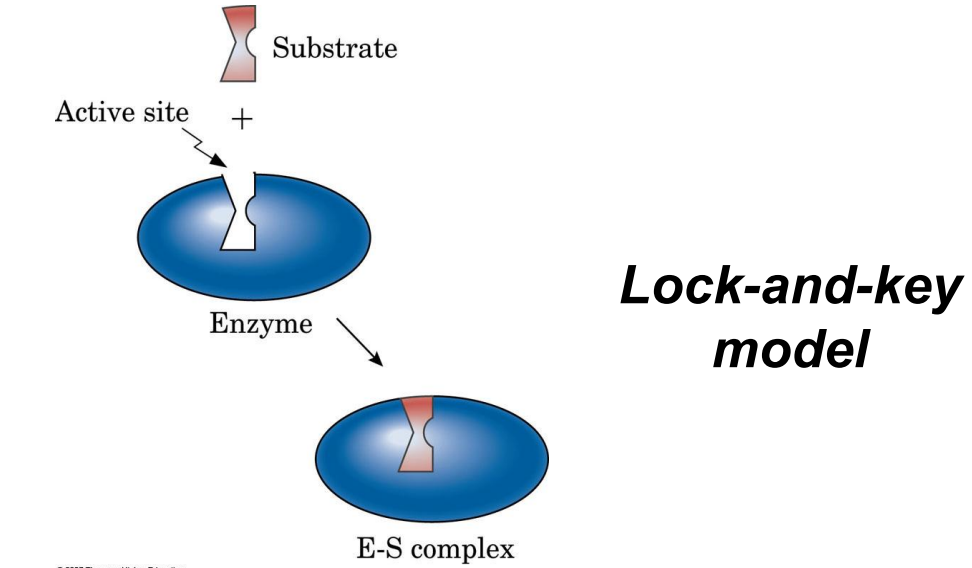
Lock-and-Key Model: Active site is a rigid shape complementary to the substrate.
Induced Fit Model: Active site molds to fit the substrate upon binding, allowing greater flexibility.
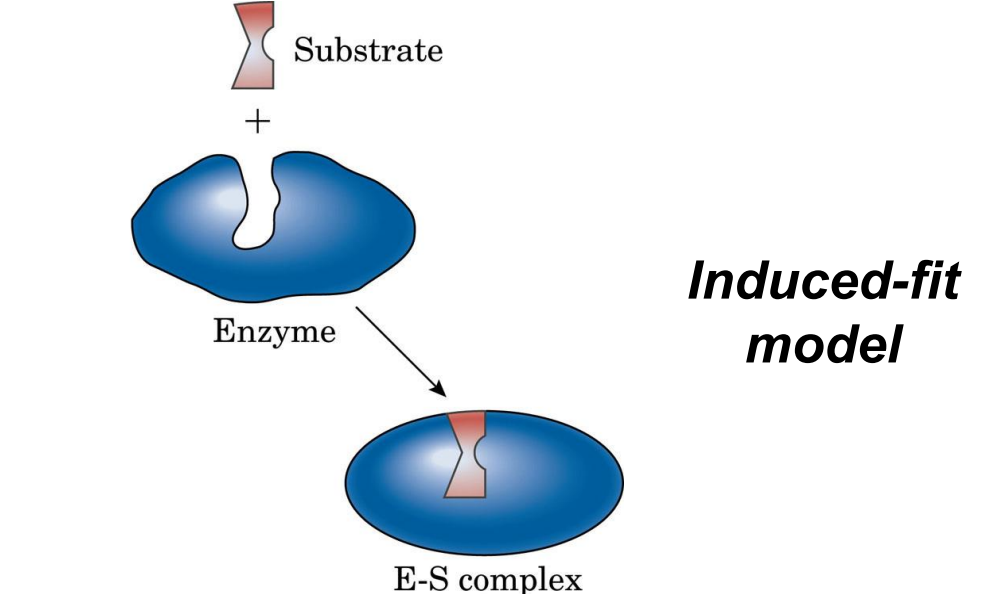
Forces in Binding: Hydrogen bonds, hydrophobic, and electrostatic interactions stabilize the enzyme-substrate complex.
Factors Affecting Enzyme Activity
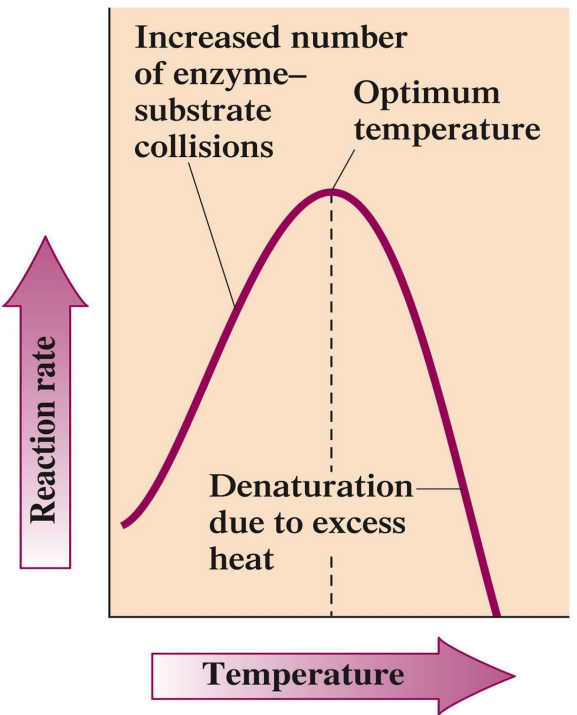
Temperature: Optimal at ~37°C for human enzymes; activity increases with temperature up to a certain point before denaturation.
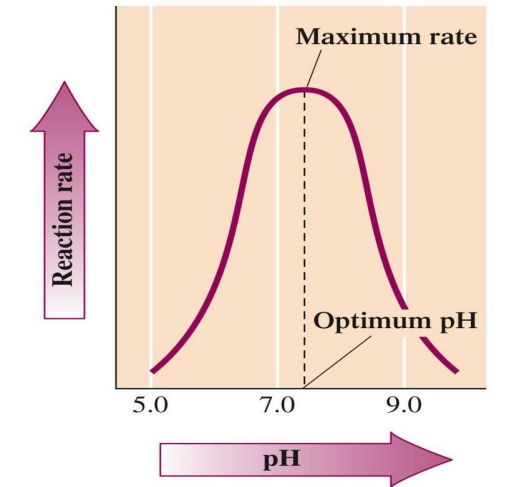
pH: Optimal pH varies; most enzymes work best around pH 7.0-7.5.
Substrate Concentration: Increases activity up to a saturation point where all active sites are occupied.
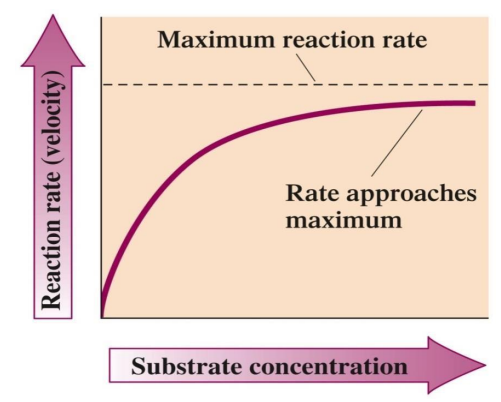
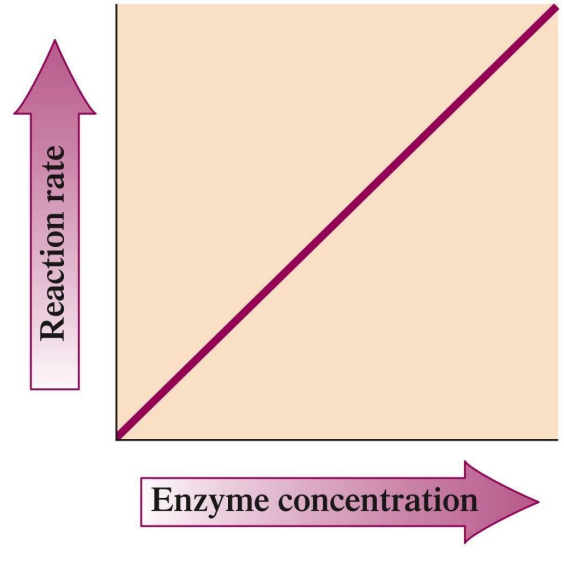
Enzyme Concentration: Higher concentrations lead to a faster reaction rate, provided sufficient substrate.
Enzyme Regulation and Inhibition
Activation: Initiation or increase in enzyme activity.
Inhibition:
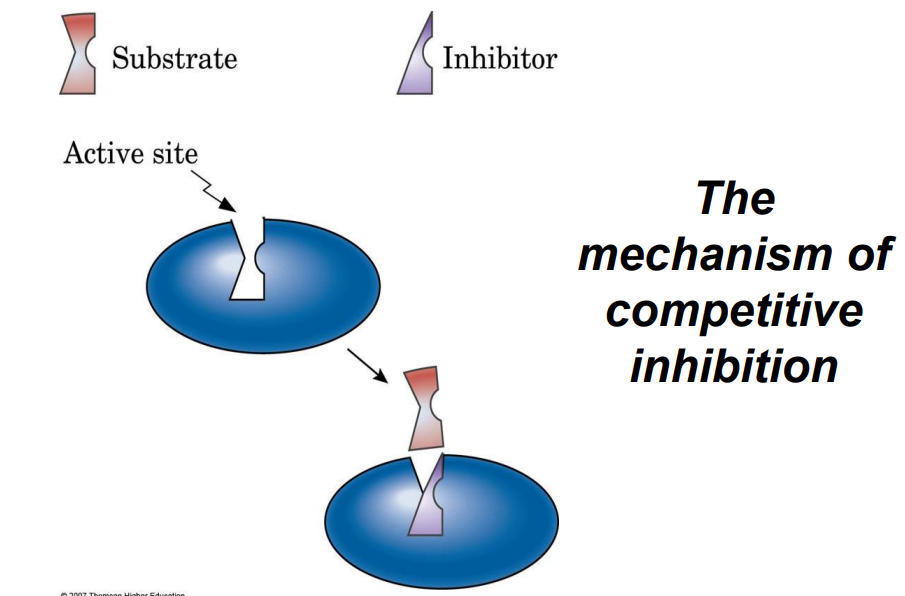
Competitive Inhibitors: Bind to the active site, competing with the substrate.
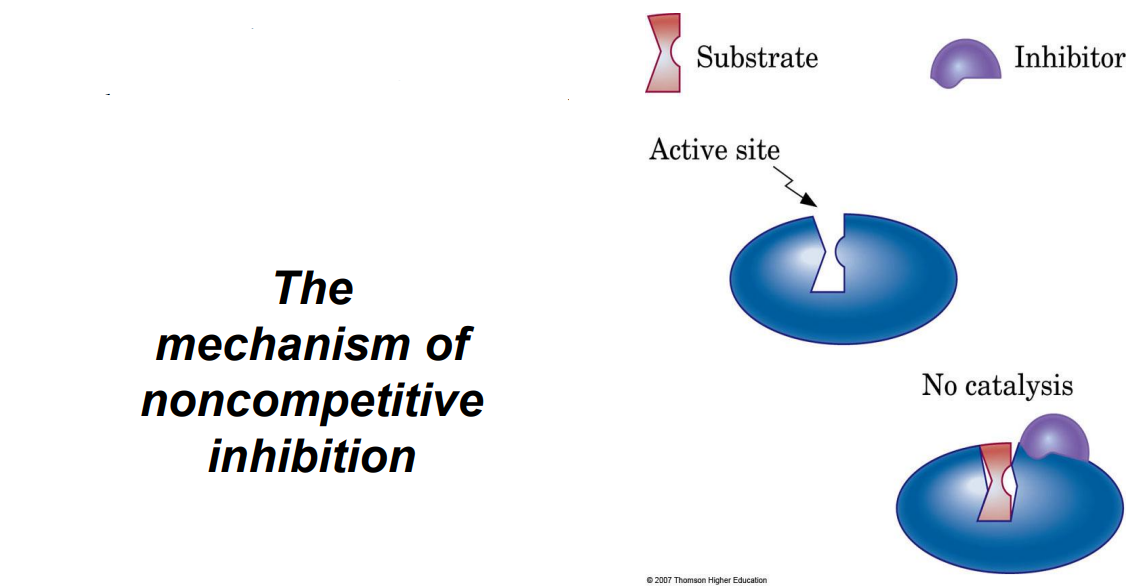
Noncompetitive Inhibitors: Bind elsewhere, altering the enzyme’s shape so it can’t bind the substrate.
Irreversible Inhibition: Covalent binding to the active site, permanently inactivating the enzyme (e.g., nerve agents).
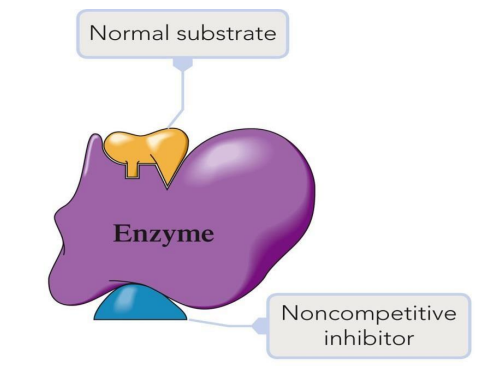
Enzyme Regulation Mechanisms
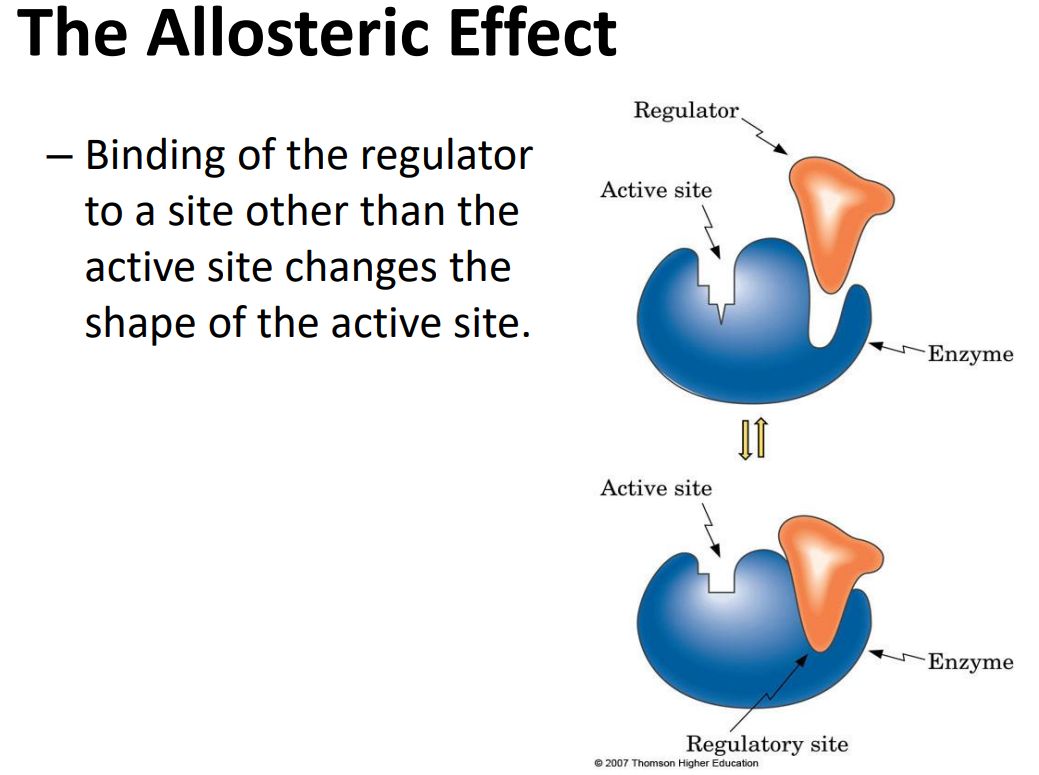
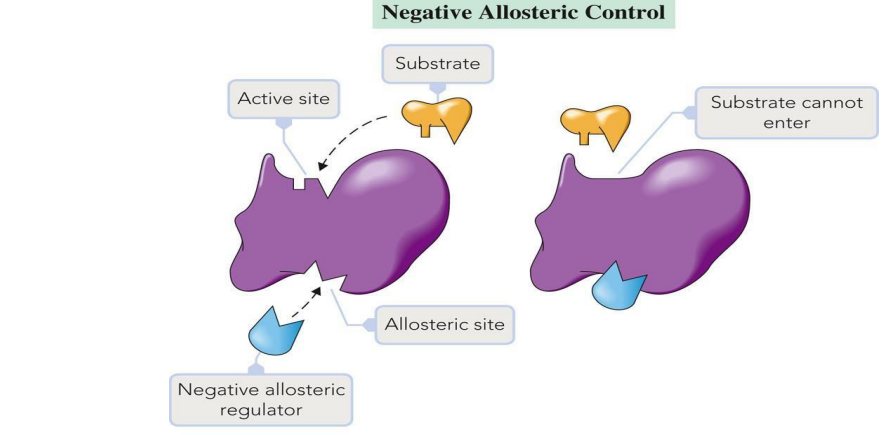
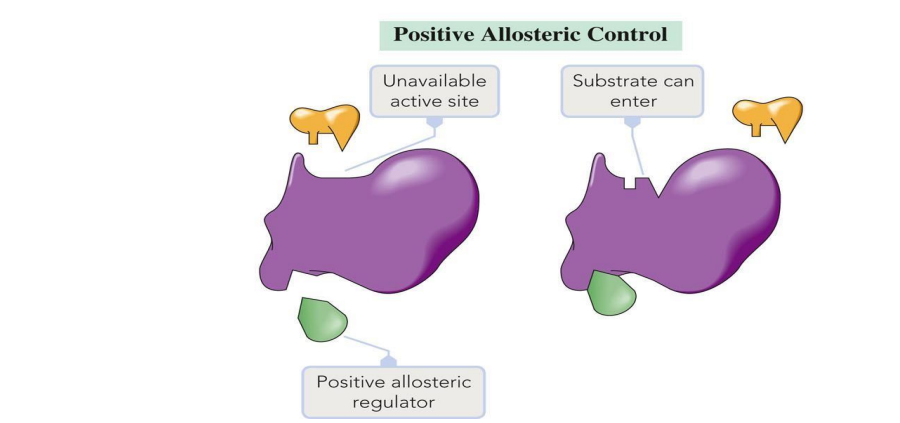
Allosteric Regulation: Molecules bind at sites other than the active site to regulate activity.
Negative Modulation: Inhibition.
Positive Modulation: Activation.
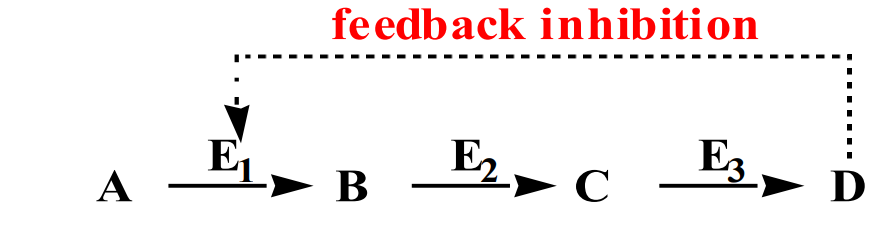
Feedback Control: End-product of a reaction pathway inhibits an enzyme in an earlier step to prevent excess product formation.
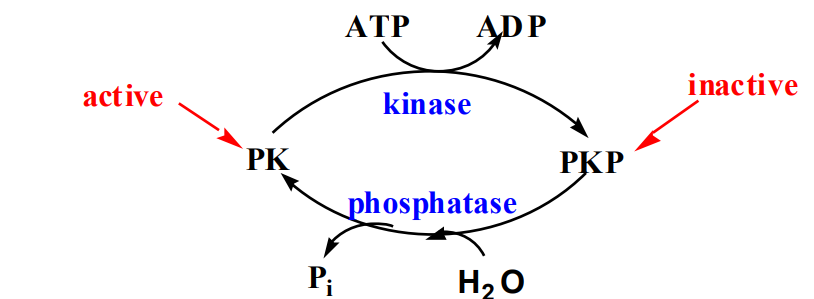
Protein Modification: Covalent modification, such as phosphorylation, changes enzyme activity.
Isoenzymes: Different forms of the same enzyme (e.g., LDH in heart and liver), providing tissue-specific activity.
Clinical Applications of Enzymes
Enzyme levels in blood can indicate tissue damage (e.g., high LDH for heart attack).
Diagnostic enzymes in various diseases:
ALT: Liver damage.
ALP: Liver or bone disease.
Amylase: Pancreatic disease.
LDH: Heart attack or hepatitis.
Key Points for Review
Enzymes reduce activation energy, not consumed in reactions.
Active Site: Specific, unique to each enzyme, complements substrate shape.
Inhibitors: Can be competitive (bind active site) or noncompetitive (bind elsewhere).
Optimal Conditions: Each enzyme has an optimal pH and temperature for peak activity.
Clinical Relevance: Enzymes serve as indicators in diagnostics for organ-specific damage.
PRACTICE QUESTIONS
Multiple Choice Questions (20)
What are enzymes primarily composed of?
a) Lipids
b) Carbohydrates
c) Proteins
d) Nucleic acids
Answer: c) Proteins
Explanation: Enzymes are mostly large, globular proteins.
What is the role of a cofactor in enzyme activity?
a) It destroys enzymes
b) It is a substrate
c) It is necessary for activity
d) It is the enzyme product
Answer: c) It is necessary for activity
Explanation: Cofactors are nonprotein components necessary for enzyme function.
Which of the following suffixes do most enzymes have?
a) -ase
b) -in
c) -ose
d) -peptide
Answer: a) -ase
Explanation: Most enzymes are named with the suffix "-ase."
Which enzyme class catalyzes hydrolysis reactions?
a) Transferases
b) Hydrolyases
c) Ligases
d) Isomerases
Answer: b) Hydrolyases
Explanation: Hydrolyases catalyze reactions that add water to break bonds.
What model describes how the enzyme active site is complementary to the substrate shape?
a) Induced Fit Model
b) Lock-and-Key Model
c) Catalytic Model
d) Substrate-Affinity Model
Answer: b) Lock-and-Key Model
Explanation: This model states that the active site is a rigid shape that fits the substrate.
Which of the following is NOT a type of enzyme regulation?
a) Allosteric regulation
b) Feedback inhibition
c) Competitive inhibition
d) Passive regulation
Answer: d) Passive regulation
Explanation: There is no such regulation termed "passive."
What is the active site of an enzyme?
a) The site where products are formed
b) The part where the substrate binds
c) The area where cofactors bind
d) The location of enzyme denaturation
Answer: b) The part where the substrate binds
Explanation: The active site is where the substrate binds and catalysis occurs.
Competitive inhibitors act by:
a) Altering the enzyme shape
b) Binding to the active site
c) Binding to the cofactor
d) Increasing substrate concentration
Answer: b) Binding to the active site
Explanation: Competitive inhibitors compete with the substrate for the active site.
Which reaction type do ligases catalyze?
a) Hydrolysis
b) Bond formation
c) Isomerization
d) Electron transfer
Answer: b) Bond formation
Explanation: Ligases catalyze the joining of two molecules, often using ATP.
What effect does temperature have on enzyme activity?
a) Always increases activity
b) Optimal at ~37°C but can denature above
c) Has no effect
d) Decreases activity at all times
Answer: b) Optimal at ~37°C but can denature above
Explanation: Enzymes have an optimal temperature, after which they can denature.
Which class of enzymes would catalyze the following reaction: AB + C → A + BC?
a) Ligases
b) Isomerases
c) Lyases
d) Oxidoreductases
Answer: c) Lyases
Explanation: Lyases add or remove groups to form or break double bonds.
Which enzyme subclass transfers phosphate groups?
a) Kinases
b) Peroxidases
c) Hydrolases
d) Decarboxylases
Answer: a) Kinases
Explanation: Kinases transfer phosphate groups often from ATP.
What term describes enzymes that are inhibited by their own products?
a) Allosteric enzymes
b) Feedback inhibitors
c) Competitive inhibitors
d) Irreversible inhibitors
Answer: b) Feedback inhibitors
Explanation: Feedback inhibition occurs when the end product inhibits an earlier enzyme in the pathway.
Which type of enzyme acts specifically on the bond between two amino acids?
a) Glycosidases
b) Esterases
c) Proteases
d) Kinases
Answer: c) Proteases
Explanation: Proteases break peptide bonds between amino acids.
Which of the following is an example of a coenzyme?
a) Zn²⁺
b) NAD⁺
c) H₂O
d) ATP
Answer: b) NAD⁺
Explanation: NAD⁺ is an organic cofactor that assists in enzyme function.
Which enzyme class includes those that transfer hydrogen atoms?
a) Ligases
b) Oxidoreductases
c) Transferases
d) Lyases
Answer: b) Oxidoreductases
Explanation: Oxidoreductases facilitate oxidation-reduction reactions.
Which of the following statements about enzyme specificity is true?
a) All enzymes have absolute specificity
b) Enzymes can only act on one substrate
c) Enzymes can have varying specificities
d) All enzymes can work on any substrate
Answer: c) Enzymes can have varying specificities
Explanation: Enzymes can either have absolute specificity or a broader specificity depending on their type.
What happens to an enzyme at very high pH levels?
a) It becomes more active
b) It is unaffected
c) It denatures
d) It converts into a coenzyme
Answer: c) It denatures
Explanation: High pH can lead to denaturation of proteins, including enzymes.
Which of the following processes does NOT involve enzymes?
a) DNA replication
b) Metabolism
c) Passive diffusion
d) Protein synthesis
Answer: c) Passive diffusion
Explanation: Passive diffusion does not require enzymes.
Which model allows for flexibility of the active site to better fit the substrate upon binding?
a) Lock-and-Key Model
b) Induced Fit Model
c) Enzyme-Substrate Complex Model
d) Rigid Structure Model
Answer: b) Induced Fit Model
Explanation: The induced fit model suggests the active site molds to fit the substrate more effectively after initial contact.
Identification Questions (10)
Apoenzyme: The protein part of an enzyme that is inactive on its own.
Cofactor: A nonprotein component that is necessary for enzyme activity (e.g., metal ions like Zn²⁺).
Coenzyme: An organic cofactor, often a vitamin, that assists in enzyme function.
Active Site: The specific part of an enzyme where the substrate binds and catalysis occurs.
Catalytic Site: The location within an enzyme where the actual chemical reaction takes place.
Feedback Control: A regulatory mechanism where the end product of a pathway inhibits an enzyme in an earlier step, preventing overproduction.
Isoenzymes: Different forms of the same enzyme that catalyze the same reaction but are found in different tissues and may have different kinetics.
Allosteric Regulation: A form of regulation where molecules bind to sites other than the active site to enhance or inhibit enzyme activity.
Saturated Solution: A point where increasing substrate concentration no longer increases reaction rate because all active sites are occupied.
Competitive Inhibition: A type of inhibition where the inhibitor competes with the substrate for binding to the active site.
Essay Questions (2)
Discuss the differences between competitive and noncompetitive inhibition. Include examples and the impact each type of inhibition has on enzyme function.
Answer: Competitive inhibition occurs when an inhibitor molecule competes with the substrate for binding to the enzyme's active site. This type of inhibition can be overcome by increasing substrate concentration. An example of a competitive inhibitor is the drug methotrexate, which inhibits the enzyme dihydrofolate reductase by competing with its substrate. Noncompetitive inhibition, on the other hand, occurs when the inhibitor binds to an enzyme at a site other than the active site, causing a change in enzyme shape that reduces activity regardless of substrate concentration. An example is heavy metals like lead that can bind to enzymes and inhibit their function. Both types of inhibition can affect enzyme kinetics and reduce the rate of product formation but in different ways.
Explain the importance of enzyme specificity and how it relates to enzyme function in biological systems. Describe how factors such as pH and temperature influence this specificity and activity.
Answer: Enzyme specificity is crucial as it ensures that enzymes catalyze only specific reactions, allowing for tight regulation of metabolic pathways in biological systems. Specificity is determined by the enzyme's active site, which matches only specific substrates, akin to a lock and key. Factors such as pH and temperature significantly influence enzyme activity. Each enzyme has an optimal pH range (usually around 7.0-7.5 for many enzymes) and a temperature range (usually optimal at around 37°C for human enzymes) where it functions best. Outside these optimal ranges, enzymes can denature or lose their shape, leading to decreased specificity and reaction rates, potentially affecting overall metabolism and biochemical processes.
NUCLEIC ACIDS
Nucleic Acids
Structure of the Nucleotide
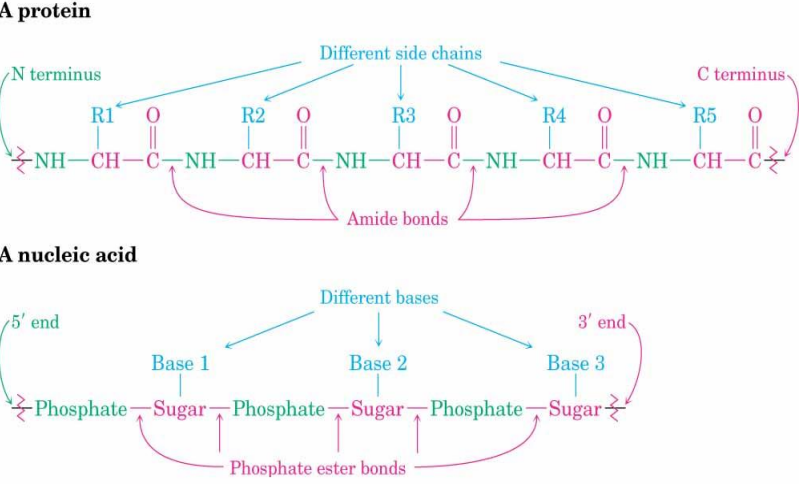
DNA and RNA are polymers whose monomer units are called nucleotides.
A nucleotide itself consists of:A nitrogen-containing heterocyclic base.
A ribose or deoxyribose sugar ring.
A phosphoric acid unit.
DNA and RNA
Deoxyribonucleic acid (DNA) and ribonucleic acid (RNA) are the chemical carriers of genetic information.
Nucleic acids are biopolymers made of nucleotides, aldopentoses linked to a purine or pyrimidine and a phosphate.
Sugars in DNA and RNA
RNA is derived from ribose.
DNA is derived from 2′-deoxyribose (the
'is used to refer to positions on the sugar portion of a nucleotide).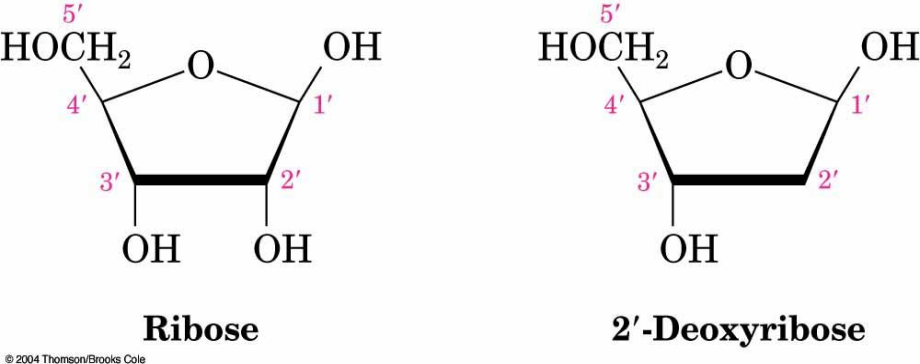
Nucleotides
A nucleotide is the repeating unit of the DNA or RNA polymer.
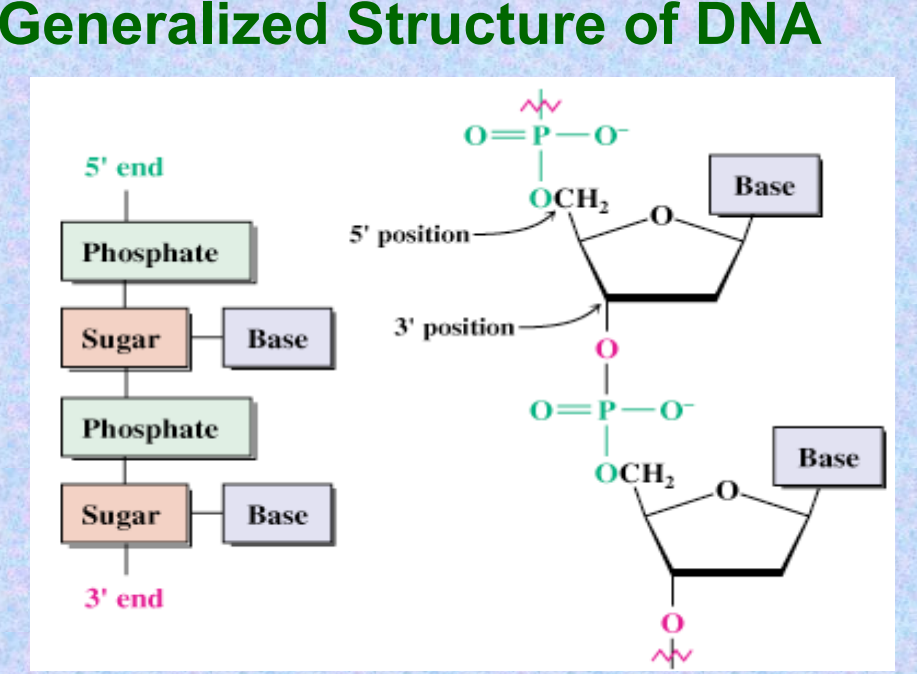
In DNA and RNA, the base is bonded to C1′ of the sugar, and the phosphate is bonded to C5′ (and connected to 3′ of the next unit).
Naming Nucleotides
Begin with the name of the nitrogenous base.
Remove the -ine ending and replace with:
-osine for purines or -idine for pyrimidines. Uracil: -acil with -idine.
If ribose: then ribonucleotide.
If deoxyribose: then deoxyribonucleotide.
Prefix "deoxy" is used before the base name for deoxyribonucleotides.
Add prefix for the number of phosphoryl groups:
Monophosphate, diphosphate, triphosphate.
Generalized Structure of DNA
DNA-Secondary Structure
The most common form of DNA is the β form. Its structure was determined by Watson and Crick in 1953.
This DNA consists of two chains of nucleotides coiled around one another in a right-handed double helix.
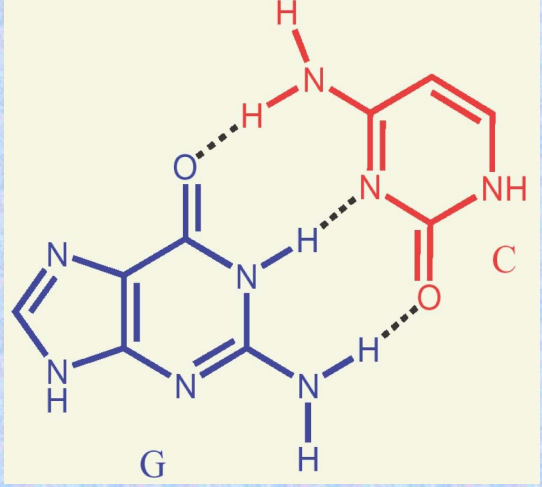
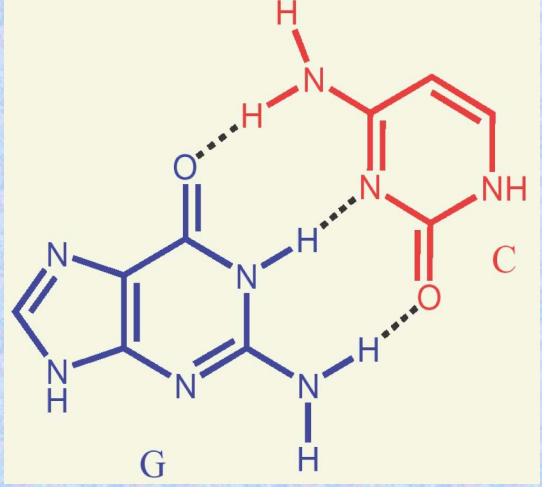
The chains run antiparallel and are held together by hydrogen bonding between complementary base pairs:
A = T, G ≡ C.
H-Bonding in DNA Structure
Hydrogen bonding between A and T or G and C helps hold the chains in the double helix.
The strands are said to be complementary.
H-Bonds in DNA
The G-C base pair involves three H-bonds.
The A-T base pair involves two H-bonds.
Grooves in DNA
The strands of the DNA double helix create two continuous grooves (major and minor).
The sugar–phosphate backbone runs along the outside of the helix, and the amine bases hydrogen bond to one another on the inside.
The major groove is slightly deeper than the minor groove, and both are lined by potential hydrogen bond donors and acceptors.
B DNA Structure
Outside diameter: 2 nm.
Length of one turn of the helix: 3.4 nm and contains 10 base pairs.
Interior diameter: 1.1 nm.
Nucleic Acid Sequences
Differences arise from the sequence of bases on the individual nucleotides.
Describing a Sequence
The chain is described from the 5′ end, identifying the bases in order of occurrence.
Use abbreviations:
A for adenosine.
G for guanosine.
C for cytidine.
T for thymine (U for uracil in RNA).
A typical sequence is written as TAGGCT.
Chromosomes
Chromosomes are pieces of DNA that contain the genetic instructions, or genes, of an organism.
Prokaryotes:
Single chromosome, no true nucleus.
Chromosome is a circular DNA molecule that is supercoiled (helix is coiled on itself).
At approximately 40 sites, a complex of proteins is attached, forming a series of loops.
This structure is called the nucleoid.
Eukaryotes:
Number and size of chromosomes vary.
True nucleus with membrane-bound organelles that separate cellular functions.
DNA is organized in nucleosomes: a strand of DNA wrapped around a disk of histone proteins.
Larger structures include the 30 nm fiber and the 200 nm fiber.
RNA Structure
The sugar-phosphate backbone for ribonucleotides is linked by 3′-5′ phosphodiester bonds.
RNA molecules are usually single-stranded.
Ribose replaces deoxyribose, and uracil replaces thymine.
Base pairing between U and A and G and C results in portions of the single strand becoming double-stranded.
Nucleic Acids and Heredity
Processes in the transfer of genetic information:
Replication: Identical copies of DNA are made.
Transcription: Genetic messages are read and carried out of the cell nucleus to the ribosomes, where protein synthesis occurs.
Translation: Genetic messages are decoded to make proteins.
DNA Replication
DNA in the chromosomes replicates itself during every cell division to maintain correct genetic information.
Two strands of DNA unwind.
Each strand acts as a template.
New bases pair with their complementary bases.
Two double helices form, which are copies of the original DNA.
Overview of DNA Replication
DNA unwinds to expose the bases:
G pairs with C, A pairs with T.
DNA copies with complementary base pairs.
Two identical copies of the original DNA strand are formed.
Summary: DNA Replication
Opening up the superstructure:
During replication, condensed chromosomal structures open via signal transduction.
Acetylation and deacetylation of key lysine residues weaken DNA-histone interactions by removing a positive charge.
Relaxation of higher structures:
Topoisomerases (gyrases) relax supercoiled DNA by introducing breaks.
Broken ends are joined once supercoiling is relaxed.
Unwinding the DNA double helix:
Helicases attach to DNA strands, catalyze ATP hydrolysis, and promote the movement needed for separation of strands.
Primer/Primases:
Primers (4-15 nucleotides long) initiate daughter strand synthesis.
Primases synthesize primers and are placed approximately every 50 nucleotides on the lagging strand.
DNA Polymerases:
Key enzymes that line up nucleotides in the proper order.
On the lagging strand, DNA polymerases synthesize short fragments called Okazaki fragments.
DNA ligase joins Okazaki fragments and remaining nicks.
Mutation and Repair
Mutations: Mistakes introduced into the DNA sequence of an organism.
Point Mutation: Substitution of a single nucleotide.
Deletion: Loss of one or more nucleotides.
Insertion: Addition of one or more nucleotides.
Many mutagens are also carcinogens, which can cause cancer.
UV Damage and DNA Repair
UV light causes the formation of pyrimidine dimers on DNA strands.
Failure to repair these defects can lead to xeroderma pigmentosum, a genetic skin disorder causing sensitivity to UV light and skin cancers.
Not all mutations are harmful; some can enhance the survival rate of species.
DNA Repair
Base Excision Repair (BER):
DNA glycosylase recognizes damaged bases and hydrolyzes the β-N-glycosidic bond to excise them.
The sugar-phosphate backbone remains intact until further steps remove and replace the damaged site.
Nucleotide Excision Repair (NER):
Removes and repairs up to 24-32 nucleotides using repair enzymes.
Classes of RNA Structure
Transfer RNA (tRNA):
Transfers amino acids to the site of protein synthesis (ribosomes).
Contains the anticodon for recognizing specific codons on mRNA.
Ribosomal RNA (rRNA):
Combines with proteins to form ribosomes.
Messenger RNA (mRNA):
Directs the amino acid sequence of proteins.
Acts as a complementary copy of a gene.
Contains codons specifying amino acids.
tRNA
At least one tRNA (often several) exists for each amino acid incorporated into a protein.
Single-stranded with approximately 80 nucleotides.
Intrachain hydrogen bonding (A = U, G ≡ C) forms stems, giving the tRNA an α-helix shape.
Overall structure resembles a cloverleaf in a 3D L-shaped conformation.
Summary: Transcription
Transcription: The process by which genetic information in DNA is copied into an mRNA molecule.
DNA double helix begins to unwind near the gene to be transcribed.
Only one strand of DNA is transcribed.
Ribonucleotides assemble along the unwound DNA strand in a complementary sequence.
RNA polymerases catalyze transcription:
Poly I for rRNA formation.
Poly II for mRNA formation.
Poly III for tRNA formation.
Eukaryotic Gene Structure
Structural Gene: Transcribed into RNA; composed of:
Exons: Coding sequences.
Introns: Non-coding sequences.
Regulatory Gene: Controls transcription but is not transcribed.
Contains control elements, including the promoter.
Promoter:
Unique to each gene and contains sequences like the TATA box for initiating transcription.
Lies approximately 25 base pairs upstream of the transcription site.
Transcription Process
Initiation:
RNA polymerase binds to the promoter with the help of transcription factors.
Elongation:
RNA polymerase zips up complementary bases in the 5′ to 3′ direction.
Termination:
RNA polymerase reaches a termination sequence and transcription stops.
Post-Transcriptional Modifications
Transcribed mRNA is capped at both ends:
5′ Cap: Addition of a methylated guanine.
3′ Poly-A Tail: Addition of 100-200 adenine residues.
Splicing: Introns are removed, and exons are joined.
Similar modifications occur in tRNA and rRNA to produce functional RNA molecules.
The Genetic Code
Genetic messages in DNA are translated into mRNA:
Degenerate: More than one codon can code for the same amino acid.
Specific: Each codon specifies one amino acid.
Non-overlapping and commaless: No bases are shared between codons, and there are no noncoding bases.
Universal: The code is the same across most organisms.
64 Codons:
61 Codons code for amino acids.
3 Codons are stop signals.
Example:
CUU, CUC, CUA, CUG code for leucine (5′ → 3′ sequence).
Gene Regulation
Gene regulation controls which genes are expressed and when.
Transcriptional Level: Regulation occurs during DNA → RNA.
Involves promoters, enhancers, and response elements.
Translational Level: Regulation occurs during mRNA → Protein.
Promoters
Located adjacent to the transcription start site.
Include conserved sequences like TATA boxes.
Bind transcription factors that determine the rate of mRNA synthesis.
Enhancers
Speed up transcription.
May be several thousand nucleotides away but can loop to interact with the initiation site.
Response Elements
Activated by transcription factors in response to external stimuli like heat shock, heavy metals, or hormones.
Recombinant DNA Technology
Uses restriction enzymes to cut DNA at specific nucleotide sequences.
Steps:
Donor and plasmid DNA are cleaved by the same restriction enzyme.
Donor DNA fragment joins plasmid DNA via hydrogen bonding.
DNA ligase restores the plasmid ring.
Engineered plasmid is introduced into a bacterium for reproduction.
Polymerase Chain Reaction (PCR)
DNA amplification process involving:
Taq Polymerase: Heat-stable DNA polymerase.
Primers: Short sequences complementary to the target DNA.
Four nucleotide triphosphates.
Thermocycler: Cycles through temperatures for denaturation, annealing, and elongation.
Cycle Steps:
Denaturation: DNA strands are separated at 94–96°C.
Annealing: Primers hybridize to target sequences at 50–56°C.
Elongation: Taq polymerase synthesizes DNA at 72°C.
DNA doubles with each cycle (e.g., 1 → 2 → 4 → 8 → etc.).
PRACTICE QUESTIONS
Multiple Choice Questions (20 items)
(Each question has one correct answer with an explanation.)
What is the repeating unit of DNA or RNA polymer?
A. Ribose
B. Deoxyribose
C. Nucleotide
D. Phosphate
Answer: C. Nucleotide
Explanation: Nucleotides are the monomers that form the polymers DNA and RNA. They consist of a sugar, a base, and a phosphate group.Which sugar is found in RNA?
A. Deoxyribose
B. Fructose
C. Ribose
D. Glucose
Answer: C. Ribose
Explanation: Ribose is the sugar in RNA, whereas deoxyribose is found in DNA.In the DNA double helix, which base pairs with guanine (G)?
A. Thymine (T)
B. Adenine (A)
C. Uracil (U)
D. Cytosine (C)
Answer: D. Cytosine (C)
Explanation: Guanine pairs with cytosine through three hydrogen bonds in DNA.What is the diameter of the DNA double helix?
A. 1 nm
B. 2 nm
C. 3.4 nm
D. 10 nm
Answer: B. 2 nm
Explanation: The DNA double helix has a diameter of 2 nm.Which type of RNA carries amino acids to the ribosome?
A. mRNA
B. tRNA
C. rRNA
D. snRNA
Answer: B. tRNA
Explanation: tRNA (transfer RNA) transports amino acids to the ribosome for protein synthesis.What is the main function of RNA polymerase during transcription?
A. Unwinding the DNA
B. Synthesizing ribosomes
C. Assembling nucleotides into RNA
D. Splicing RNA
Answer: C. Assembling nucleotides into RNA
Explanation: RNA polymerase builds the RNA strand by assembling ribonucleotides complementary to the DNA template strand.What type of bonds hold base pairs together in DNA?
A. Ionic bonds
B. Covalent bonds
C. Hydrogen bonds
D. Peptide bonds
Answer: C. Hydrogen bonds
Explanation: Hydrogen bonds connect complementary base pairs (A-T and G-C) in the DNA double helix.Which base is unique to RNA?
A. Thymine
B. Uracil
C. Cytosine
D. Guanine
Answer: B. Uracil
Explanation: Uracil is present in RNA instead of thymine, which is found in DNA.What is the role of helicase in DNA replication?
A. Joining Okazaki fragments
B. Synthesizing primers
C. Unwinding the DNA double helix
D. Pairing bases
Answer: C. Unwinding the DNA double helix
Explanation: Helicase unwinds the DNA to prepare it for replication.Which process copies DNA into an identical strand?
A. Transcription
B. Translation
C. Replication
D. Transformation
Answer: C. Replication
Explanation: DNA replication produces two identical copies of the original DNA molecule.What type of mutation involves substituting one nucleotide for another?
A. Point mutation
B. Deletion
C. Insertion
D. Frameshift mutation
Answer: A. Point mutation
Explanation: A point mutation changes a single nucleotide in the DNA sequence.What forms the backbone of DNA?
A. Base pairs
B. Sugar-phosphate groups
C. Hydrogen bonds
D. Amino acids
Answer: B. Sugar-phosphate groups
Explanation: The sugar-phosphate backbone supports the structure of DNA and RNA.What is the sequence written from the 5′ to 3′ end called?
A. Genetic code
B. DNA helix
C. Nucleic acid sequence
D. Codon
Answer: C. Nucleic acid sequence
Explanation: Nucleic acid sequences are described starting from the 5′ end and listing the bases in order.What does the "A" in mRNA stand for?
A. Acceptor
B. Adenine
C. Acid
D. Amino
Answer: D. Amino
Explanation: mRNA stands for messenger RNA, which carries the message for amino acid assembly.What type of repair removes and replaces up to 24-32 nucleotides?
A. BER
B. NER
C. Recombinant repair
D. Ligase repair
Answer: B. NER
Explanation: Nucleotide excision repair (NER) is a mechanism to remove larger DNA lesions.What shape does tRNA take in its three-dimensional structure?
A. Double helix
B. Cloverleaf
C. Straight chain
D. L-shaped
Answer: D. L-shaped
Explanation: tRNA has a cloverleaf secondary structure but folds into an L-shape in 3D.What does the term "degenerate" mean in the genetic code?
A. Nonfunctional codons exist
B. A single codon codes for multiple amino acids
C. Multiple codons can code for the same amino acid
D. Some bases overlap between codons
Answer: C. Multiple codons can code for the same amino acid
Explanation: Degeneracy means redundancy in the genetic code.Which enzyme joins Okazaki fragments?
A. Helicase
B. Ligase
C. Polymerase
D. Topoisomerase
Answer: B. Ligase
Explanation: DNA ligase seals nicks between Okazaki fragments to complete DNA replication.Which codons signal the termination of protein synthesis?
A. AUG
B. UAA, UAG, UGA
C. GGG, CCC, AAA
D. UUU, UGC, UAC
Answer: B. UAA, UAG, UGA
Explanation: These are stop codons that terminate translation.What happens during transcription?
A. DNA is copied into mRNA.
B. mRNA is translated into proteins.
C. DNA is replicated.
D. tRNA transports amino acids.
Answer: A. DNA is copied into mRNA.
Explanation: Transcription converts DNA into messenger RNA.
Identification (10 items)
The sugar found in RNA.
Answer: RiboseThe enzyme that synthesizes RNA during transcription.
Answer: RNA polymeraseThe complementary base of adenine in DNA.
Answer: ThymineThe process by which DNA is copied into an identical strand.
Answer: ReplicationA mutation where nucleotides are added to the DNA sequence.
Answer: InsertionThe RNA molecule that directs the sequence of amino acids in a protein.
Answer: mRNAThe type of bond between the sugar and phosphate in DNA.
Answer: Phosphodiester bondThe purine bases found in nucleic acids.
Answer: Adenine and guanineThe enzyme responsible for sealing nicks in the DNA backbone.
Answer: DNA ligaseThe repeating sequence that makes up the backbone of DNA and RNA.
Answer: Sugar-phosphate
Essay Questions (5 items, 2 points each)
Explain how the structure of DNA supports its function in storing genetic information.
Answer: DNA's double helix structure allows it to store vast amounts of genetic information compactly. Complementary base pairing ensures accurate replication, while the sugar-phosphate backbone provides stability.Describe the main differences between DNA and RNA.
Answer: DNA contains deoxyribose and thymine, while RNA contains ribose and uracil. DNA is double-stranded, whereas RNA is single-stranded. DNA stores genetic information, while RNA functions in protein synthesis.Outline the steps of DNA replication and their importance.
Answer: Replication involves unwinding the DNA, pairing new nucleotides with the template strands, and forming two identical DNA molecules. This ensures genetic information is accurately passed to daughter cells.Discuss the role of transcription in gene expression.
Answer: Transcription converts DNA sequences into mRNA, which carries the genetic code to ribosomes. This is the first step in decoding genes into functional proteins.What is the significance of the genetic code being "universal" and "degenerate"?
Answer: The universal nature allows consistent genetic interpretation across species, while degeneracy provides resilience against mutations by allowing multiple codons to code for the same amino acid.
set 2
Which enzyme is responsible for unwinding the DNA helix during replication?
A. Ligase
B. Helicase
C. Polymerase
D. Primase
Answer: B. Helicase
Explanation: Helicase unwinds the DNA double helix to allow replication to begin.What is the direction of DNA synthesis?
A. 3′ to 5′
B. 5′ to 3′
C. Both directions simultaneously
D. Depends on the strand
Answer: B. 5′ to 3′
Explanation: DNA polymerase synthesizes new DNA strands only in the 5′ to 3′ direction.Which enzyme adds RNA primers during DNA replication?
A. DNA polymerase
B. Primase
C. Ligase
D. Helicase
Answer: B. Primase
Explanation: Primase synthesizes short RNA primers that provide a starting point for DNA polymerase.What are Okazaki fragments?
A. Pieces of RNA primers
B. Short DNA fragments on the lagging strand
C. The result of mutations during replication
D. Newly synthesized mRNA segments
Answer: B. Short DNA fragments on the lagging strand
Explanation: Okazaki fragments are created on the lagging strand during DNA replication.Which strand is synthesized continuously during DNA replication?
A. Leading strand
B. Lagging strand
C. Both strands
D. Neither strand
Answer: A. Leading strand
Explanation: The leading strand is synthesized continuously in the direction of the replication fork.What is the role of DNA ligase in replication?
A. Unwinding the helix
B. Synthesizing new DNA strands
C. Joining Okazaki fragments
D. Proofreading DNA
Answer: C. Joining Okazaki fragments
Explanation: DNA ligase seals nicks between Okazaki fragments, forming a continuous strand.What is the template strand for transcription?
A. The RNA strand
B. The DNA strand that runs 3′ to 5′
C. The DNA strand that runs 5′ to 3′
D. The mRNA strand
Answer: B. The DNA strand that runs 3′ to 5′
Explanation: The 3′ to 5′ DNA strand serves as the template for RNA synthesis, as RNA is synthesized 5′ to 3′.Which base is present in RNA but not in DNA?
A. Thymine
B. Adenine
C. Guanine
D. Uracil
Answer: D. Uracil
Explanation: Uracil replaces thymine in RNA.What is the process of making RNA from a DNA template called?
A. Replication
B. Translation
C. Transcription
D. Reverse transcription
Answer: C. Transcription
Explanation: Transcription synthesizes RNA using DNA as a template.Which enzyme catalyzes the formation of RNA during transcription?
A. DNA polymerase
B. RNA polymerase
C. Helicase
D. Primase
Answer: B. RNA polymerase
Explanation: RNA polymerase builds the RNA strand complementary to the DNA template.In transcription, RNA is synthesized in which direction?
A. 3′ to 5′
B. 5′ to 3′
C. Both directions simultaneously
D. Depends on the strand
Answer: B. 5′ to 3′
Explanation: RNA polymerase synthesizes RNA in the 5′ to 3′ direction, complementary to the 3′ to 5′ DNA template strand.What is the role of mRNA in translation?
A. It transfers amino acids to the ribosome.
B. It carries the genetic code to the ribosome.
C. It catalyzes peptide bond formation.
D. It forms the ribosome structure.
Answer: B. It carries the genetic code to the ribosome.
Explanation: mRNA contains codons that specify the amino acid sequence for protein synthesis.What molecule brings amino acids to the ribosome?
A. mRNA
B. rRNA
C. tRNA
D. DNA
Answer: C. tRNA
Explanation: tRNA delivers specific amino acids to the ribosome based on the codons in mRNA.What is the start codon in translation?
A. UAG
B. UGA
C. AUG
D. AAA
Answer: C. AUG
Explanation: AUG codes for methionine and serves as the start codon.What happens during the elongation stage of translation?
A. The ribosome assembles around the mRNA.
B. Amino acids are added to the growing polypeptide chain.
C. The ribosome reaches a stop codon.
D. tRNA binds to the ribosome.
Answer: B. Amino acids are added to the growing polypeptide chain.
Explanation: During elongation, the ribosome moves along the mRNA, adding amino acids.Which component of translation contains anticodons?
A. mRNA
B. tRNA
C. rRNA
D. Ribosome
Answer: B. tRNA
Explanation: tRNA molecules have anticodons that pair with codons on mRNA.Which of the following codons signals the end of translation?
A. AUG
B. UAA
C. CUG
D. GGU
Answer: B. UAA
Explanation: UAA is a stop codon that terminates translation.What is the name of the bond formed between amino acids during translation?
A. Hydrogen bond
B. Glycosidic bond
C. Peptide bond
D. Phosphodiester bond
Answer: C. Peptide bond
Explanation: Peptide bonds link amino acids to form proteins.Where does translation occur in the cell?
A. Nucleus
B. Cytoplasm
C. Mitochondria
D. Golgi apparatus
Answer: B. Cytoplasm
Explanation: Translation occurs in the cytoplasm, where ribosomes are located.What is the main purpose of translation?
A. To replicate DNA
B. To synthesize RNA
C. To produce proteins
D. To unwind DNA
Answer: C. To produce proteins
Explanation: Translation converts the genetic code in mRNA into a sequence of amino acids to form a protein.