AP Bio Unit 5 Genetics
Chapter 10 Meiosis
Vocab Terms:
Synapsis- the pairing of homologous chromosomes during prophase I
Synaptonemal complex- zipper-like structure composed of proteins connects a homologous chromosomes together tightly along their lengths during part of prophase I
Karyotype- display of a COMPLETE SET OF chromosome pairs of a cell arranged by size/shape
Recombinant chromosome- created when crossing over combines DNA from parents into a a single chromosome
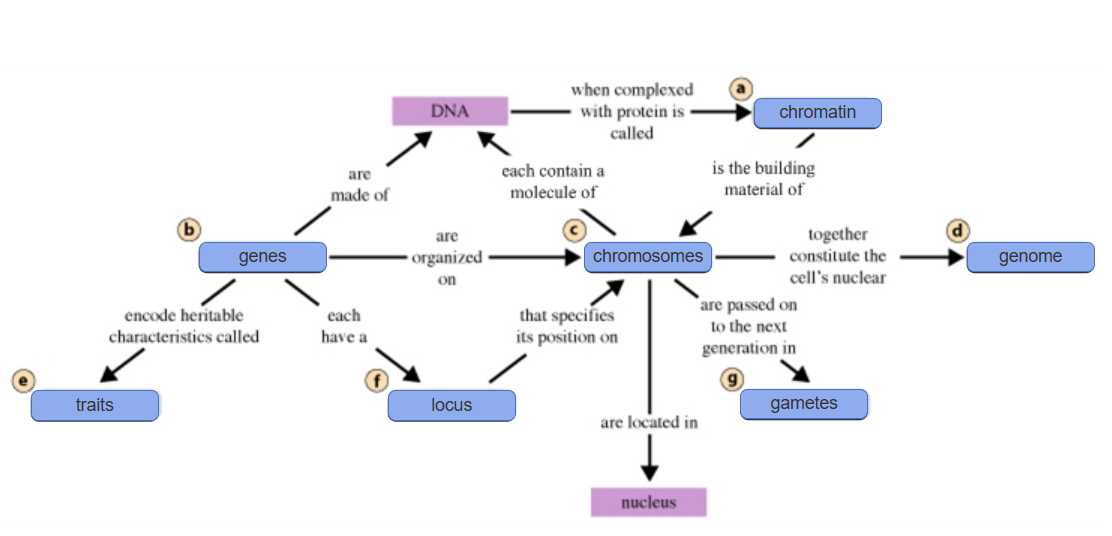
Gene- Segments of DNA that encode heritable characteristics (traits) passed on from parent to offspring
Allele- the different forms of a gene
Locus- the LOCATION of a gene on a chromosome
Tetrads- Homologous chromosome pairs
Chromosome- Thread-like structures that contain genes and carry a molecule of DNA
Homologous chromosomes- A pair of chromosomes SIMILAR in size and shape
Chromatid- A single half of a chromosome (chromosome comes in set of 2 sister chromatids)
Chromatin- the complex of DNA and proteins that serve as the BUILDING MATERIAL for chromosomes when wound up into coils
Genome- An entire (haploid) set of chromosomes found in a cell
*Sporophyte- the diploid, multicellular stage of the plant that produces haploid spores by meiosis (like gametes produced in animals plants produce spores)
Haploid (n) one set of chromosomes (23) NO HOMOLOGOUS PAIRS
Meiosis Steps
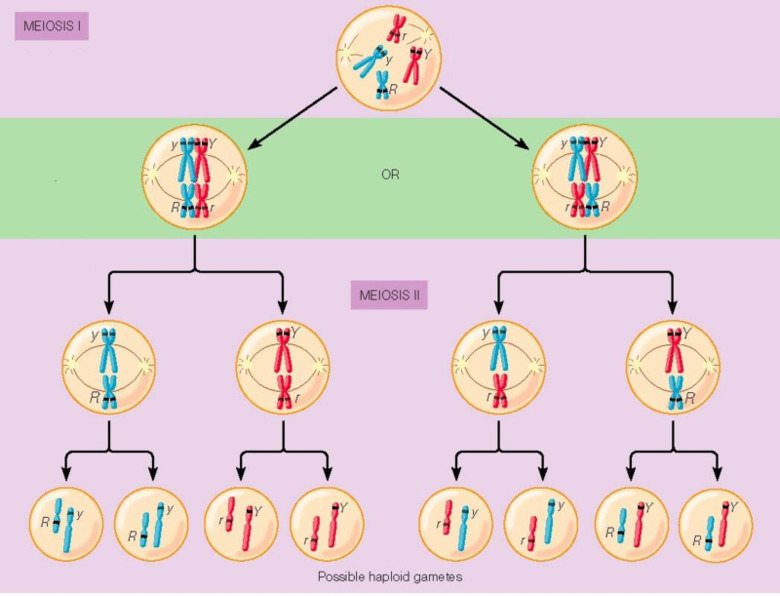
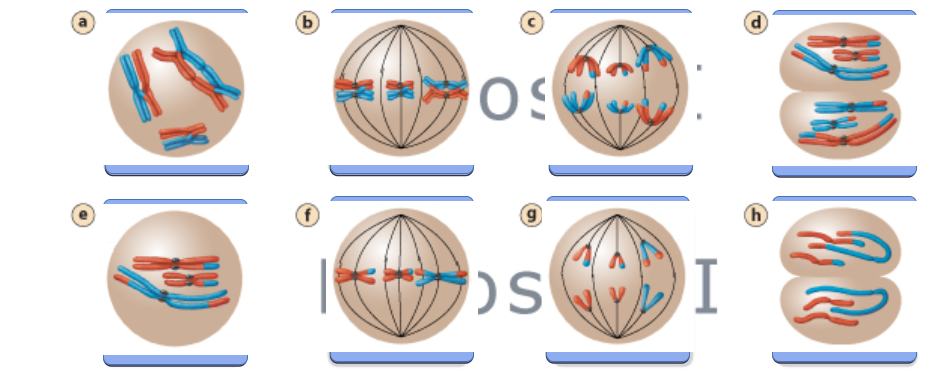
Meiosis I
This stage separates homologous chromosomes, reducing the chromosome number from diploid (2n) to haploid (n).
Prophase I
Chromosomes condense and become visible.
Homologous chromosomes pair up in a process called synapsis, forming tetrads (Homologous chromosome pairs)
Crossing over occurs at chiasmata, increasing genetic diversity.
The nuclear envelope breaks down, and spindle fibers begin to form.
Metaphase I
Homologous chromosome pairs (tetrads) align along the metaphase plate and are attached to the spindle apparatus.
Independent Assortment- random orientation of homologous pairs on the equator
Calculating possible configurations: Based on n of pairs of homologous chromosomes 2^n ex. n = 3 then there would be 8 possible configurations
NOT 2^6 because in Metaphase I separate based on homologous chromosome pairs
Spindle fibers attach to centromeres of homologous chromosomes.
Anaphase I
Homologous chromosomes are pulled apart to opposite poles of the cell.
Sister chromatids remain attached at their centromeres.
Different from mitosis where sister chromatids separate
Telophase I and Cytokinesis
Chromosomes reach the poles, and the nuclear membrane may reform.
The cell divides into two each contains HALF the original number of chromosomes Ex. 4 → 2 or 2 pairs → 1 pair
Cell divides into TWO haploid daughter cells—NO HOMOLOGOUS PAIRS because separated in prior steps
Meiosis II (Equational Division)
This stage separates sister chromatids, similar to mitosis.
Prophase II
Similar to prophase of mitosis except no DNA replication prior
Chromosomes condense again if they had decondensed.
The nuclear envelope dissolves, and spindle fibers reform.
Metaphase II
Chromosomes align at the metaphase plate, similar to mitosis.
Spindle fibers attach to centromeres (center) on each chromatid
Anaphase II
Sister chromatids are finally pulled apart to opposite poles.
Telophase II and Cytokinesis
Nuclear membranes reform around each set of chromosomes.
The cytoplasm divides, resulting in FOUR haploid daughter cells, each genetically unique containing chromatids.
Mechanisms contributing to genetic variation:
Errors (mutations) in DNA replication
Separation of sister chromatids in meiosis II
Fertilization- random fusion of male & female gametes
Independent assortment- homologous chromosomes randomly lined up during Metaphase I to be separated into gametes later

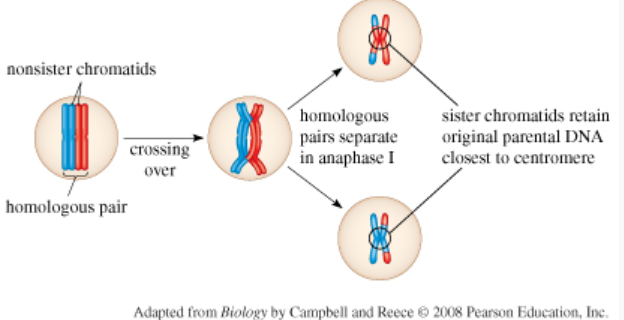
Crossing over producing new chromosome combinations
Occurs in Prophase 1 at the chiasmata main purpose is the produce genetic variation
Also important for synapsis to occur correctly—matching of homologous chromosome pairs to ensure each cell receives one complete set of chromosomes
Typically occurs at the ends of chromosomes—retain DNA near centromere
Homologous chromosomes must align precisely so that non-sister chromatids can exchange corresponding segments of DNA
As a result of crossing over, sister chromatids are no longer identical
Meiosis I | Meiosis II |
DNA Content (# of copies of chromosomes) is cut in HALF Ploidy level- (# of complete SETS of chromosomes) changes from diploid (46) to haploid (32); separation of homologous chromosomes decreases the ploidy level from 2n to n and produces daughter cells with a single chromosome set Homologous chromosomes separate after Meiosis I—alleles also separate Sister chromatid cohesion and crossing over hold homologous chromosomes together until anaphase I where they separate. Cohesions at the arms of the chromosomes are cut for homologous chromosomes to separate | DNA Content (# of copies of chromosomes) is cut in ½ again Ploidy level- (# of complete SETS of chromosomes) remains haploid Sister chromatids separate after Meiosis II Cohesions at the centromeres (center) of chromosomes are cut for the sister chromatids to separate At the end they produce four haploid daughter cells |
Concept 10.1
Asexual reproduction occurs when a sole parent passes copies of all genes to offspring without gamete fusion → creating a clone (genetically identical individuals)
Concept 10.2 Fertilization and Meiosis alternate in sexual life cycles
Normal human somatic cells (body cells, non-sex cells) are diploid (2 sets of chromosomes) with n = 46 chromosomes, one set inherited from each parent
Diploids have 22 homologous pairs (44 chromosomes) autosomes and one pair of sex chromosomes that help determine the sex of the person (XX vs XY)
Reproductive organs produce haploid gametes through meiosis—each gamete is haploid with one set of n = 23 chromosomes
During fertilization gametes (egg and sperm) combine to form a diploid zygote with 2 sets of chromosomes n = 46 that develops into an organism via mitosis
Chapter 11 Mendelian Genetics
Vocab Terminology
Character: A heritable feature that varies among individuals (i.e. color)
Quantitative character- heritable feature that continuously varies over a range of values (can be quantified) (i.e. height, weight, blood pressure)
Each variant for a character (i.e. white flowers vs purple flowers)
*Epistasis- interaction between alleles of DIFFERENT genes where phenotypic expression of one gene alters that of another gene—one gene masks the expression of another (ex. aacc same as AAcc if c is epistatic to A)
*Pleiotropy- ability of a SINGLE gene to have multiple effects
Hybridization- crossing of two TRUE-BREEDING varieties
Generations: P generation—parents; F1 generation—first filial generations cross from parents; F2 generation—second filial generation crossing F1 generations
Polygenic inheritance: Phenotype controlled by multiple genes rather than one
Multifactoral: phenotypic character influenced by multiple genes/environmental factors
Mendel’s Model
1) Alleles/alternative versions of genes account for variations in inherited characters
Results from slight variations in nucleotide sequence along chromosomes
2) For each character, an organism inherits TWO versions of a gene, one from each
3) If two alleles in an organism differ (heterozygous), then one is a dominant allele that determines the organism’s appearance and the other recessive allele has no noticeable effect
4) Law of segregation—the two alleles segregate (separate from each other) during gamete formation and end up in different gametes; Used for monohybrid cross
Egg/sperm gets only one of the two alleles present in somatic cells of the organism (i.e. with heterozygous 50% of gametes receive dominant and 50% receive recessive allele)
In the case of the heterozygous green-pod plant (Gg), one gamete will receive the dominant allele (G), and the other gamete will receive the recessive allele (g)
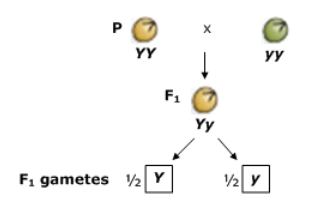
Heterozygote—has two different alleles for a gene (heterozygous); homozygote—same alleles for a gene (homozygous)
Phenotype- the appearance/observable traits (purple or white)
Genotype- the genetic makeup (PP, Pp, pp)
Testcross
Used to determine the genotype of an unknown dominant trait (whether it is homozygous or heterozygous) by crossing with a recessive
If homozygous all offspring will show the dominant trait because dominant allele is always present; if heterozygous, half of the offspring will show the recessive trait
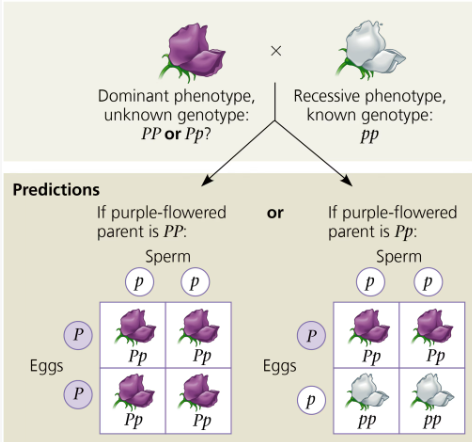
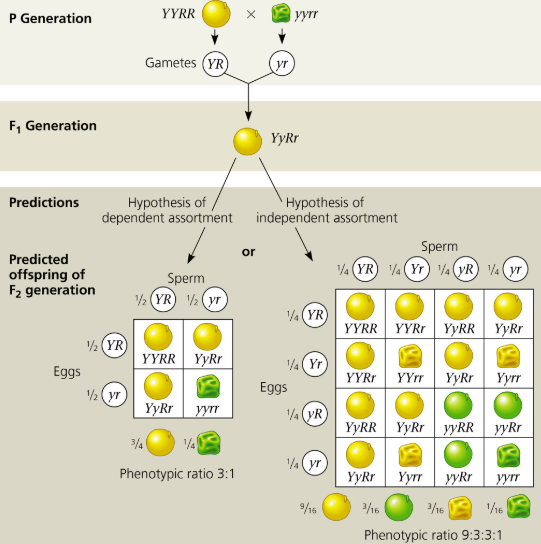
Law of Independent Assortment
Law of Independent Assortment—two or more genes assort independently—each pair of alleles segregates independently during gamete formation; Used for dihybrid cross
Law of Segregation based on a SINGLE character with all F1 generations being monohybrids—heterozygous for one particular being followed in the cross (Monohybrid cross)
Dihybrids- individual heterozygous for TWO characters being followed in the cross (Dihybrid cross)
Statistics of Inheritance—laws of probability govern mendelian inheritance
Multiplication Rule-
Probability that 2+ INDEPENDENT events will occur together in a specific combination → multiply probabilities of each event
Ex. Crossing AABbCc x AaBbCc probability of AaBbcc
Probability of each TRAIT (denoted by the different letters A, B, C ) → ½ (A) x ½ (B) x ¼ (C) = 1/16
AA x Aa = 50% chance of Aa or AA; Bb x Bb = 50% chance of Bb and 25% chance of BB or bb; Cc x Cc = 50% chance of Cc and 25% chance of cc or CC
Ex. Crossing Rr x Rr to get RR or rr
½ probability for carrying dominant allele (R) or recessive (r ) probability that you will get TWO recessive alleles present is ½ x ½ = ¼
Using Multiplication rule for Dihybrid Cross
Use monohybrid cross to find probability of each INDIVIDUAL trait
For each of the 4 possible combinations, multiply together the ratios of the respective individual traits
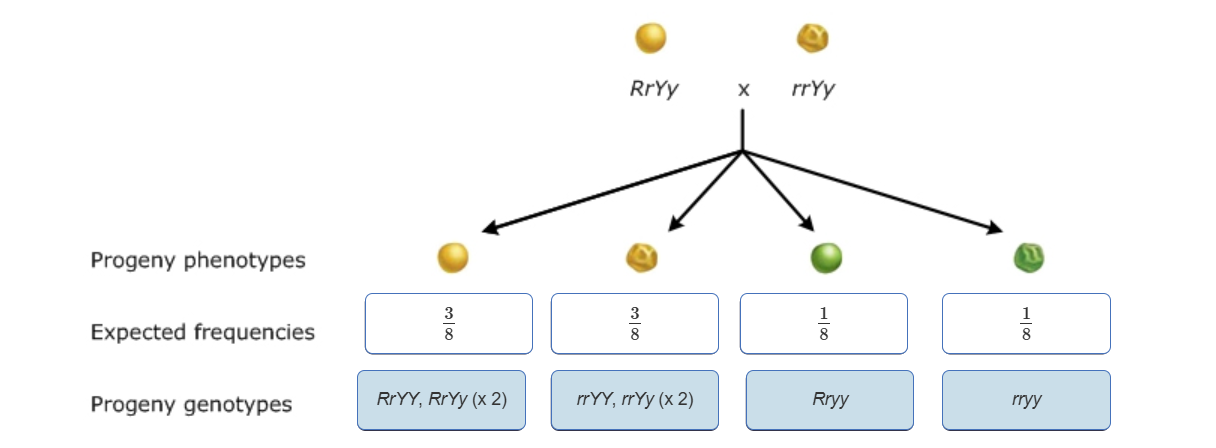
Addition Rule-
Probability that 2+ MUTUALLY EXCLUSIVE events will occur → add together individual probabilities
Ex. Throwing a die landing on a 4 OR 5 → 1/6 + 1/6 = 1/3
Ex. Crossing Rr x Rr to get Rr
½ probability of rR and ½ probability of Rr → ¼ + ¼ = ½
More Complex Genetics
Incomplete Dominance- When hybrids (heterozygous) have an appearance BETWEEN that of 2 parents (i.e. red x white = pink) (i.e. C^R and C^W)
Complete Dominance- heterozygote & homozygote for dominant allele are indistinguishable
Codominance- phenotype of BOTH alleles is expressed (i.e. red hair x white hair = roan horses)
Indicated not by capital/lowercase but by exponent (i.e. L^M vs L^N)
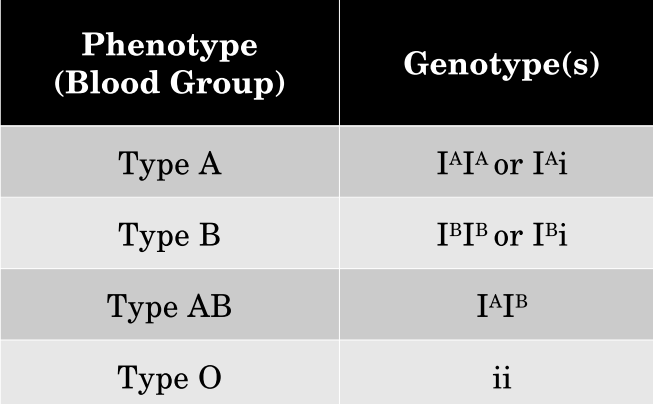
Multiple Alleles- Gene has 2+ alleles for the trait
Ex. Human ABO blood groups
I^A, I^B, i: I^A & I^B are Codominant creating type AB blood; i is recessive creating type O blood
Chi-Squared (X²) test
Used to determine if there is a significant difference between the expected and observed data
Null Hypothesis- NO statistically significant different between expected and observed data
Formula:
X² = the sum of (O - E)²/E; Observed frequencies, Expected frequencies
Steps:
Determine the null hypothesis
Use formula to calculate the X² value
n = number of categories, e = expected frequency/value, o= observed frequency/value
Calculate expected frequency/count—multiple total by expected percentage to get numbers
Plug into formula (observed value - expected value)²/expected value
Add all together for X² value
Find df with number of categories - 1
Find critical value using table (Use p = 0.05 (default) or p = 0.01)
P-value probability- how often our results could happen due to change
Degrees of freedom (df) = n - 1
If X² < critical value…FAIL to reject the null hypothesis
Differences in data due to change
If X² > critical value…reject the null hypothesis
Differences in data NOT due to chance
Example
Total M&M’s—100; 6 types of M&M’s each color has 20; Claimed percentage - 14% yellow
Expected frequencies- Total * (Percent/100)
Calculation:
(O-E)²/E = (20 - 14)²/14 = 2.57…. add all M&M’s together to get SUM(X²) = 21.01
df = n - 1 = 6 - 1 = 5
p = 0.05
critical value = 11.07 (look on table using p value and degrees of freedom)
RESULT: 21.01 > 11.07 we REJECT the null hypothesis because X² > critical value
General Rules
Crossing 2 heterozygous Xx & Xx:
Phenotype: 3:1 ratio— ¾ dominant phenotype and ¼ recessive phenotype
Genotype: 50% chance of heterozygous (Xx) 25% chance of homozygous dominant, 25% chance of homozygous recessive (XX or xx)
Crossing homozygous and heterozygous:
Phenotype: 1:1 ratio. ½ chance of heterozygous (Xx) ½ of homozygous same as homozygous parent alleles (either dominant or recessive)
Heterozygous Dihybrid cross PpRr x PpRr
Phenotype: 9:3:3:1; 9 Dominant trait for both (Pp/PP + Rr/RR) 3 Dominant/recessive trait (Pp/PP + rr) 3 Dominant/recessive trait (pp + Rr/rr) + 1 recessive for both (pprr) 16 total
Heterozygous x Homozygous Dihybrid cross PpRr x pprr:
Phenotype: 1:1:1:1 ratio 4/16 4/16 Dominant trait for both (Pp/PP + Rr/RR) 4/16 Dominant/recessive trait (Pp/PP + rr) 4/16 Dominant/recessive trait (pp + Rr/rr) + 4/16 recessive for both (pprr) 16 total
Parent genotypes from progeny
Assuming (R) is smooth (r ) is wrinkled; (Y) is yellow (y) is green
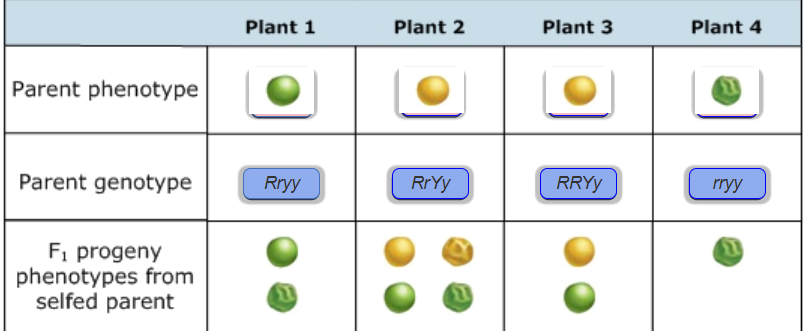
Punnett Square Predictions
Given the parent combination is Yy x Yy and yellow (Y) is dominant

Sex-Linked Chromosomes
Sex chromosomes carry sex-linked genes (MAJORITY ON THE X CHROMOSOME) males who inherit a recessive sex-linked
Sex-linked gene: Located on X or Y chromosome
Barr body- Inactive X chromosome; regulate gene dosage in FEMALES during embryonic development (when there is XX)
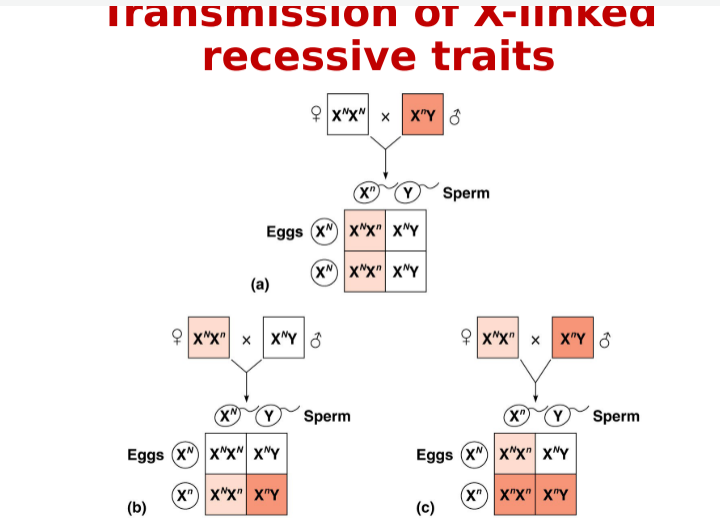
Linkage Maps
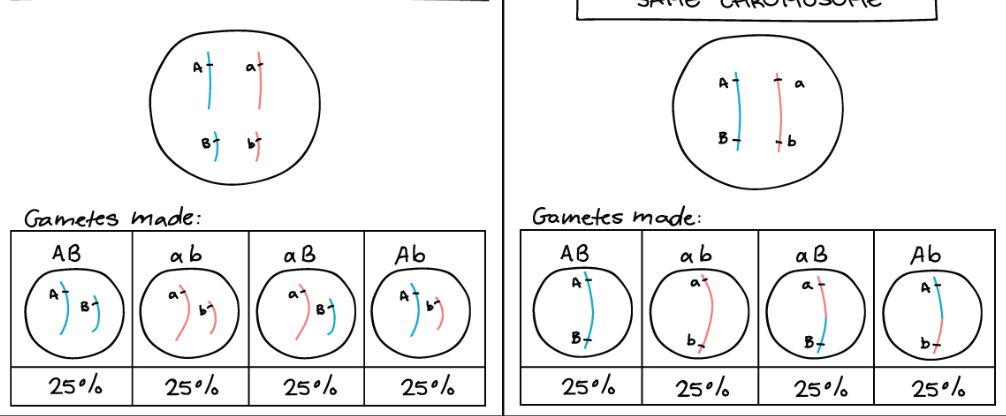
Genes on different chromosomes/far apart assort independently and are UNLINKED
Genes closer together are LINKED- alleles together on one chromosome inherited as a unit often
Recombination frequency- used to check if two genes are linked
1 map unit = 1% recombination frequency
50% recombination = FAR APART on the same chromosome/2 different chromosomes
Usually genes assort independently (allele received for one doesnt affect another) in a double heterozygous (AaBb) results in formation of 4 types of gametes with equal ¼ frequency (Combine the A alleles with the B alleles)
This is caused by the random orientation of homologous chromosome pairs during meiosis—independent assortment
Genes CLOSE TOGETHER (linked) DO NOT ASSORT INDEPENDENTLY they tend to stick together during meiosis
Most likely to contain parental configurations (alleles already together on the chromosome prior to meiosis) rarely contain recombinant configuration (results from crossing over)
DO NOT follow Mendel’s Law of Independent Assortment
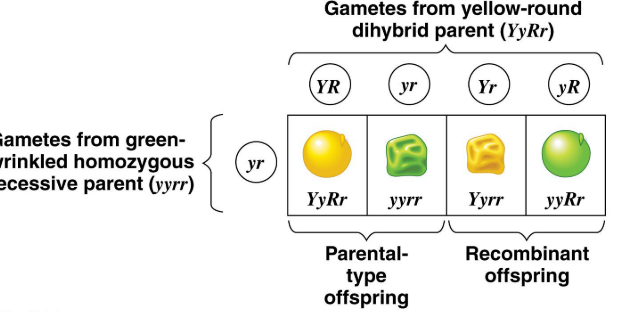
Gene Recombination
Production of offspring with combinations of traits different from either parent; Similar to parent → Parental; Different from parents → Recombinant
Finding recombinant frequency = # of Recombinants/total # offspring x 100%
MAX recombinant frequency percentage is 50%
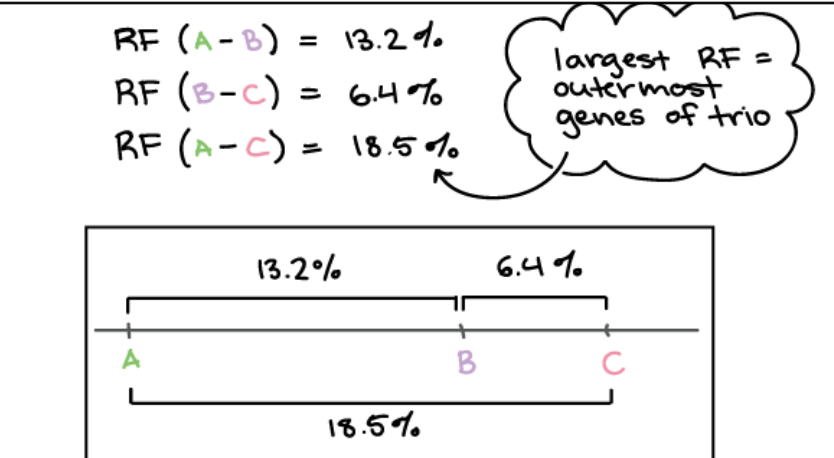
Using recombinant frequency to find order of genes
Ex A, B, C genes divide into three possible pairs (AC, AB, BC) to figure out which ones lie furthest apart → pair of genes with the largest recombination frequency must flank the third gene