L14 trafficking 1
Protein Synthesis and Ribosomes
Proteins are synthesized by ribosomes
Typical mammalian cells contain up to 10,000 different proteins.
Majority synthesized by free cytosolic ribosomes.
1/3 of proteins are synthesized by ribosomes on the endoplasmic reticulum (ER) membrane.
Synthesis of Proteins on Membrane-Bound vs. Free Ribosomes
Types of Ribosomes
1/3rd of the human proteome is synthesized on the rough ER.
Produces:
Secreted proteins
Integral membrane proteins
Soluble proteins of organelles
Proteins synthesized on free ribosomes:
Cytosolic proteins
Cytosolic peripheral membrane proteins
Nuclear proteins
Proteins targeted to mitochondria, chloroplasts, and peroxisomes.
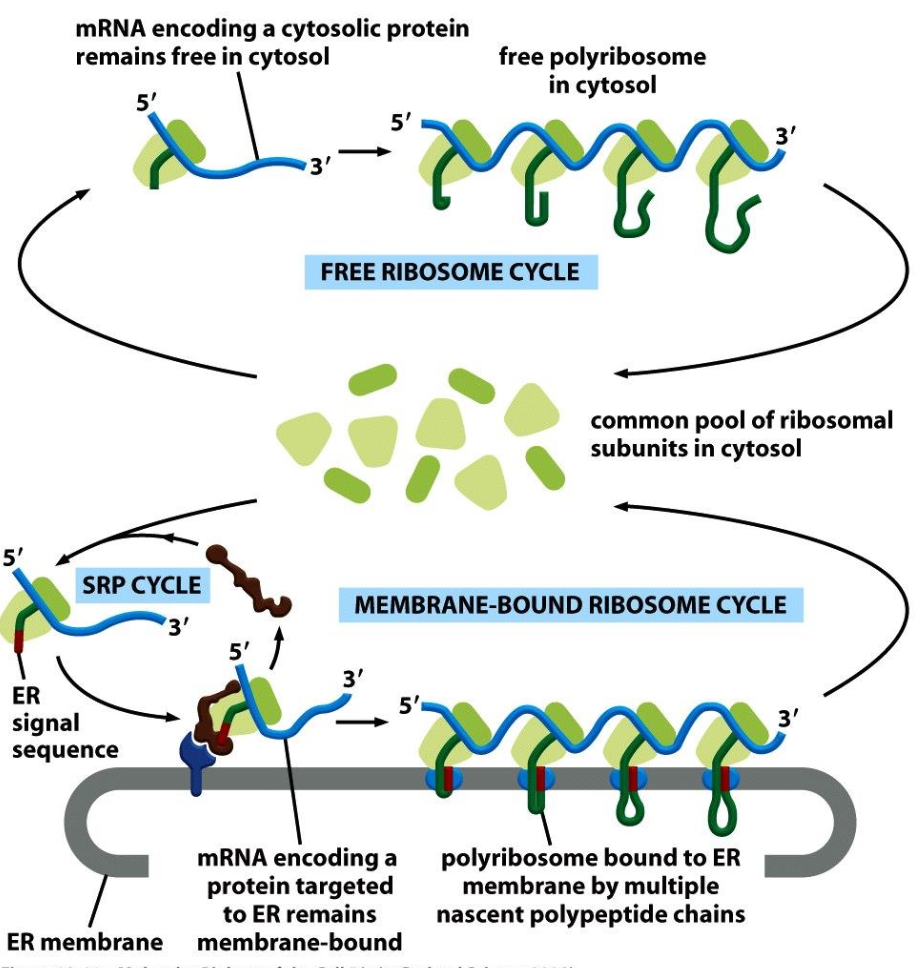
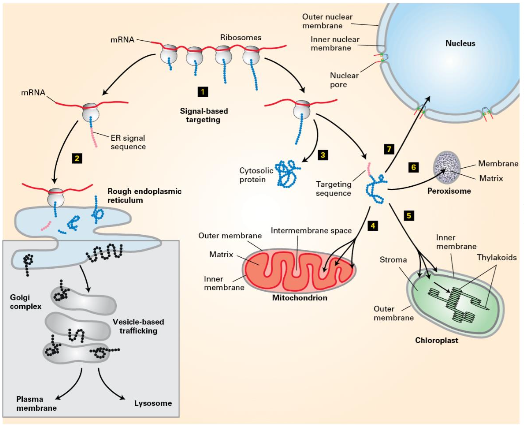
Protein Sorting Mechanisms
Key Components
Outer nuclear membrane
mRNA
Ribosomes
Inner nuclear membrane with nuclear pore
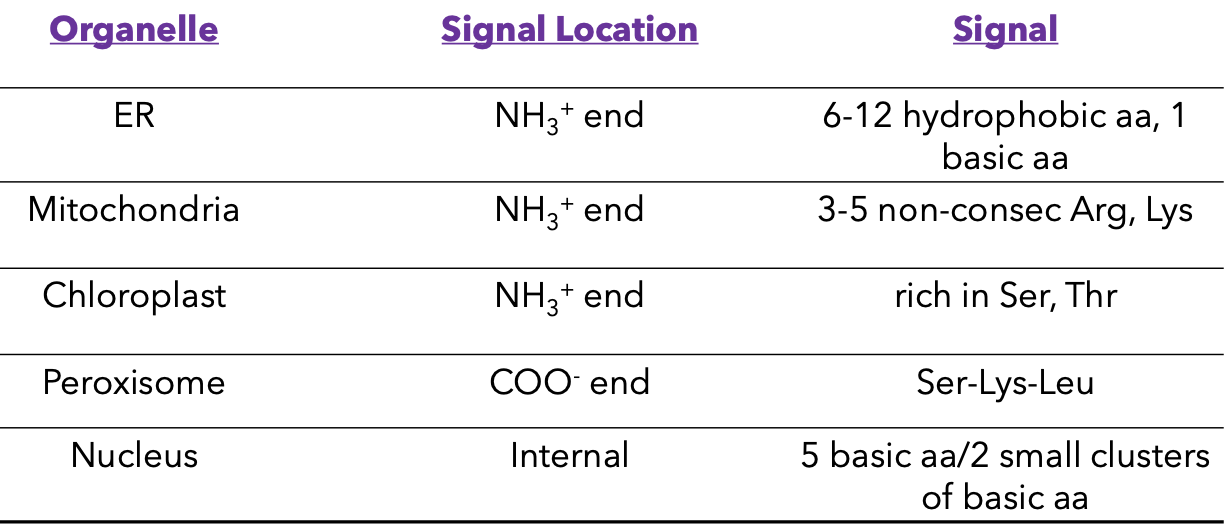
Signal-based targeting involves various sequences.
Rough ER sequence
Peroxisome sequence
Vesicle-based trafficking mechanisms illustrated.
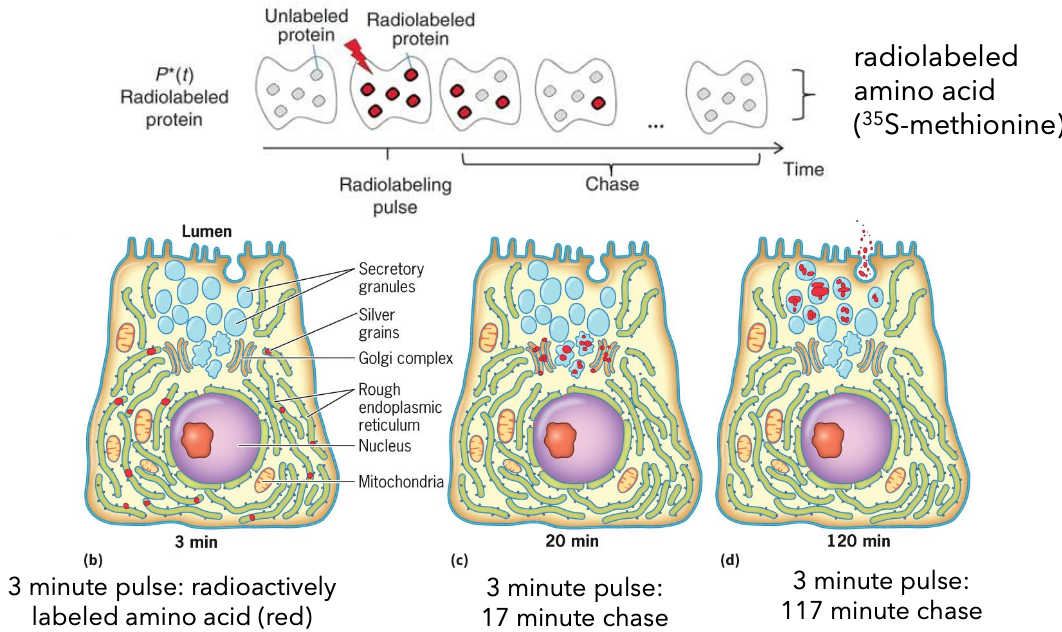
Experimental Methods to Study Protein Sorting/Transport
Pulse Chase Experiment: This technique involves labeling newly synthesized proteins with a radioactive or fluorescent tag, followed by a chase period where the label is removed, allowing researchers to track the movement and processing of these proteins over time.
Radiolabeled amino acid: 35S-methionine
Pulse phase (3 minutes): Radioactively labeled amino acid.
Chase phase: 17-minute and 117-minute chases to observe transport.
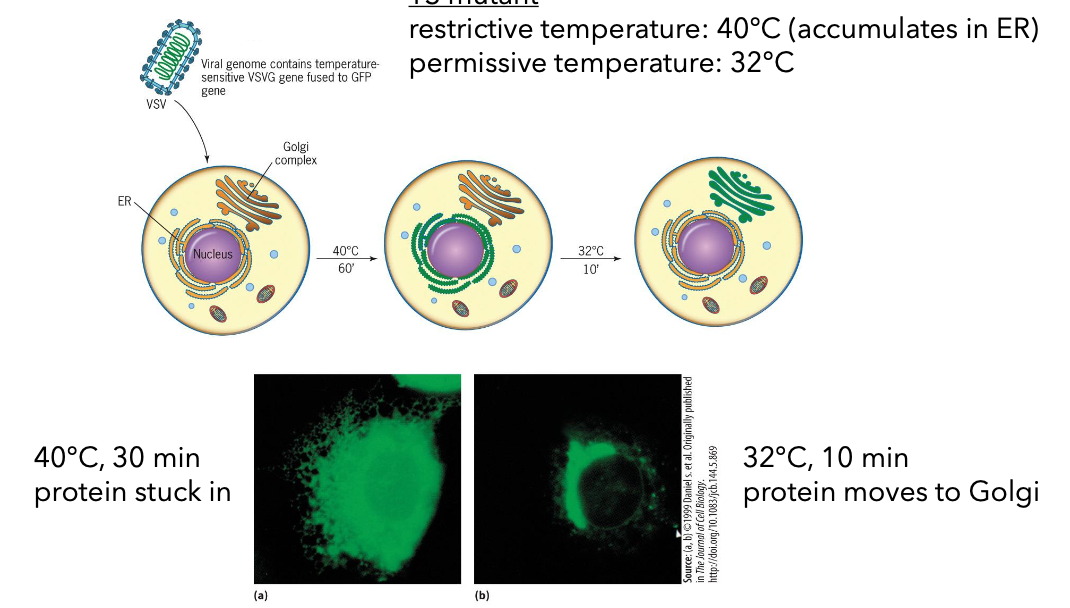
GFP-based Protein Tracking
Temperatures influence protein tracking:
Restrictive temperature (40°C): Protein accumulates in ER.
Permissive temperature (32°C): Protein moves to Golgi
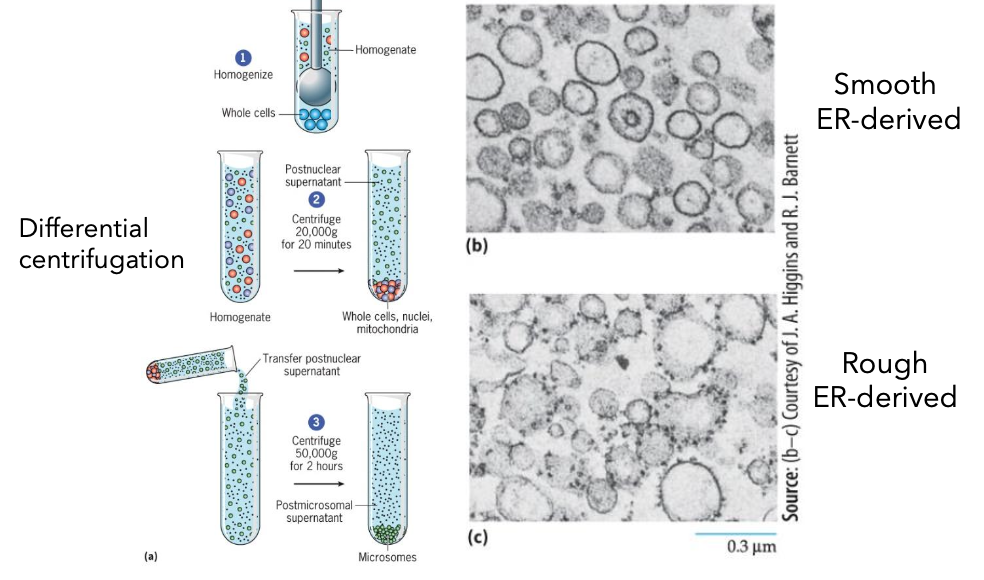
Subcellular Fractionation
Homogenize cells into a whole cells homogenate.
Differential centrifugation (20,000g for 20 minutes) yields postnuclear supernatant.
Another centrifugation (50,000g for 2 hours) creates post-microsomal supernatant containing fractions.
Analysis of Genetic Mutants
Nobel Prize 2013: Research on Saccharomyces cerevisiae (budding yeast).
sec12: Shows vesicle formation at the ER with expanded ER.
sec17: Causes accumulation of vesicles in the cell.
Page 11: Protein Sorting/Protein Targeting
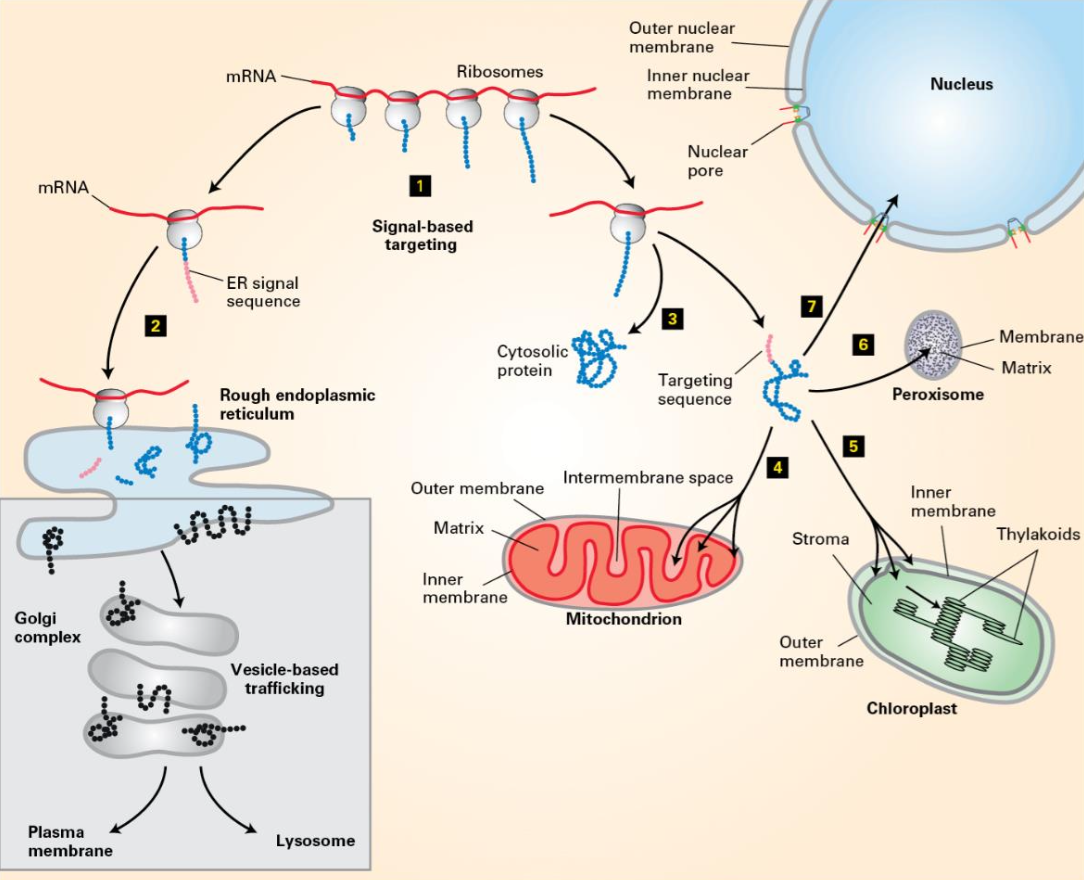
Signal-based targeting directly targets newly synthesized proteins to organelles.
Vesicle-based targeting (secretory pathway) noted.
Sorting Signals
Signal sequence
Encoded in amino acid sequence or attached carbohydrates.
Recognized by receptors:
For soluble proteins: Receptors can be integral membrane proteins.
For transmembrane proteins: Receptors are coat components.
Targeting can occur during or after protein synthesis.
Organelle Signal Locations
Signaling mechanisms for targeting proteins:
Signal Sequences Examples
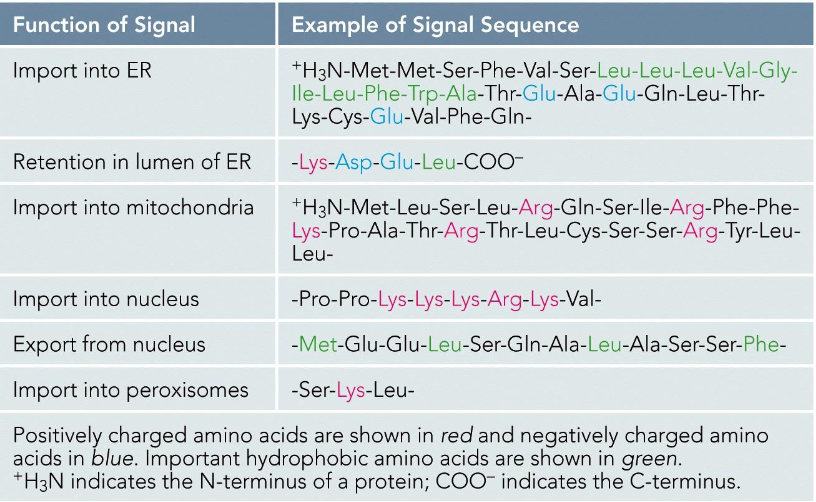
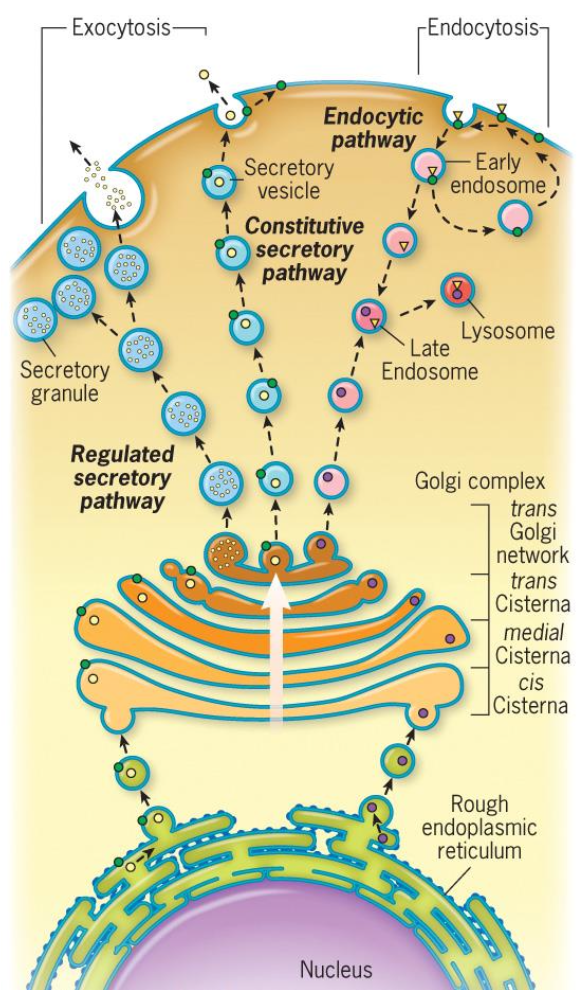
Vesicle based trafficking
Endomembrane system include: ER, Golgi complex, endosomes, lysosomes, vacuoles, seretory vesicles and granules
Biosynthetic pathway: Synthesis, modification, and transport of proteins.
Secretory pathway: where proteins are discharged (secreted) fro the cell.
Constitutive secretion: in a Continuous fashion. important for forming PM and extracellular matrix.
Regulated secretion: In response to stimuli, important for cellular signaling.
Overview of Vesicle based trafficking
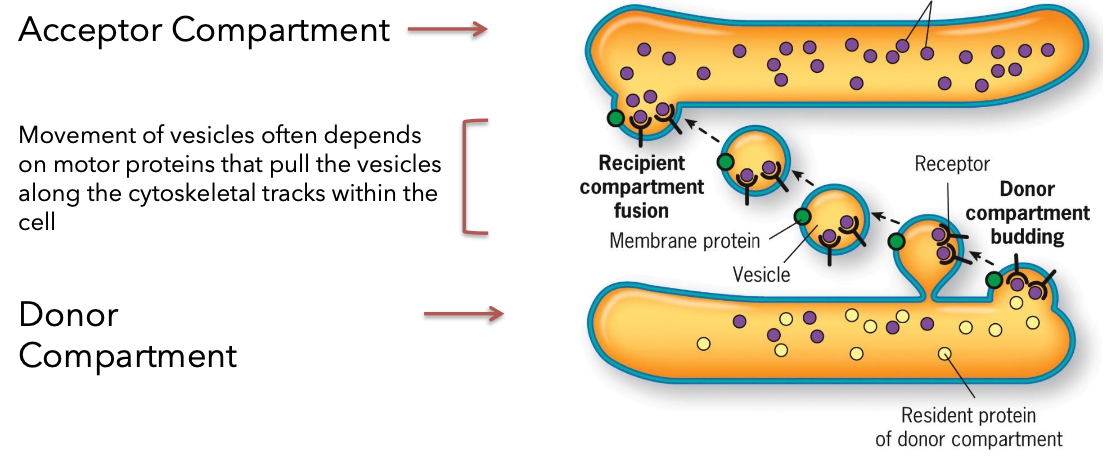
Endoplasmic Reticulum Structure
Types: Rough ER (RER) and Smooth ER (SER).
Differences between SER and RER in function and structure in conjunction with Golgi apparatus.
The Smooth ER
synthesis of steroid hormones in endocrine cells
detoxification in the liver of various organic compounds
sequetration of calcium ion from cytoplasm of muscle cells (has high conc. of calcium binding proteins)
Leydig cell example: Extensive SER for steroid hormone synthesis.
The Rough ER
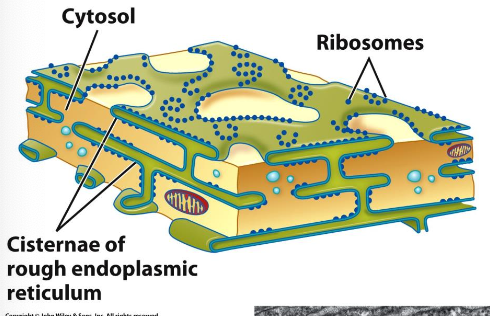
Immunofluorescence techniques used to visualize rough ER.
Continuous with outer membrane of the nuclear envelope.
Serves as the starting point for the biosynthetic/secretory pathway.
Membrane Biosynthesis in the ER
Membranes arise from pre-existing membranes.
Most lipids synthesized in ER, which are asymmetric facing cytosol and luminal/ extracellular face .
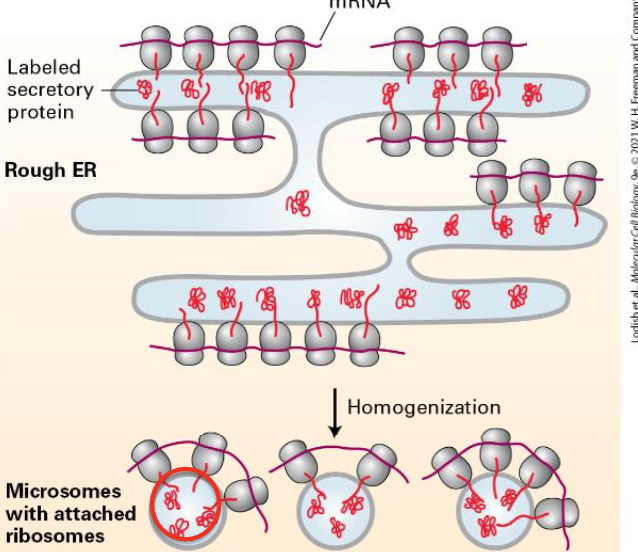
Evidence of Secretory Proteins in ER
The Pulse Chase Experiemnt: radiolabeled amino acids
Newly synthesized proteins radioactively labeled.
Homogenizing the cell leads to small vesicles called microsomes.
Ribosomes found on the outside of microsomes.
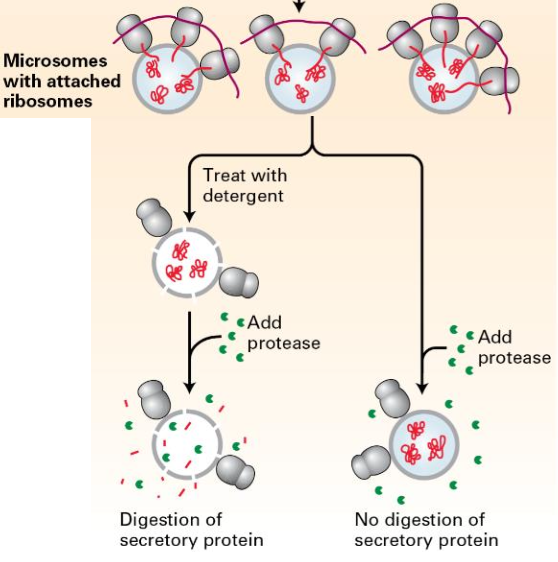
Protease experiment:
Sample 1: Add protease, Proteins inside the microsomes protected.
Sample 2: Detergent added before protease; secretory proteins degraded.
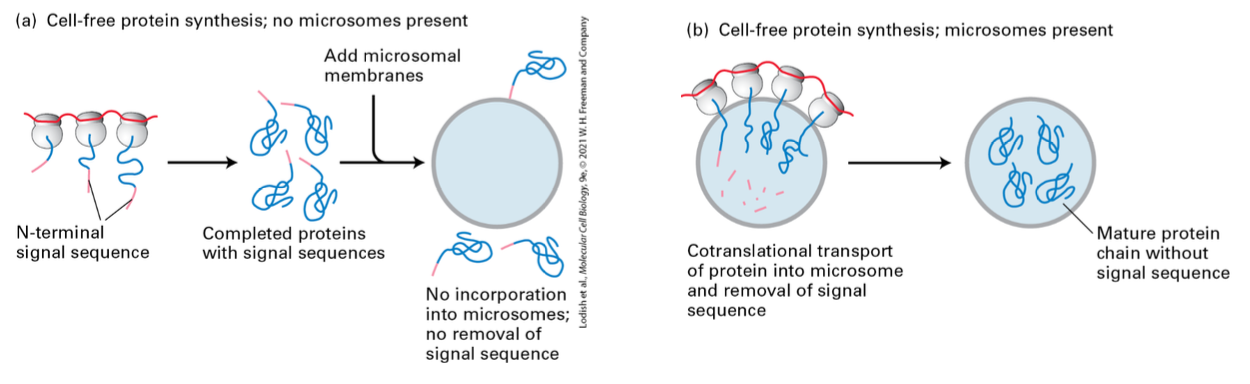
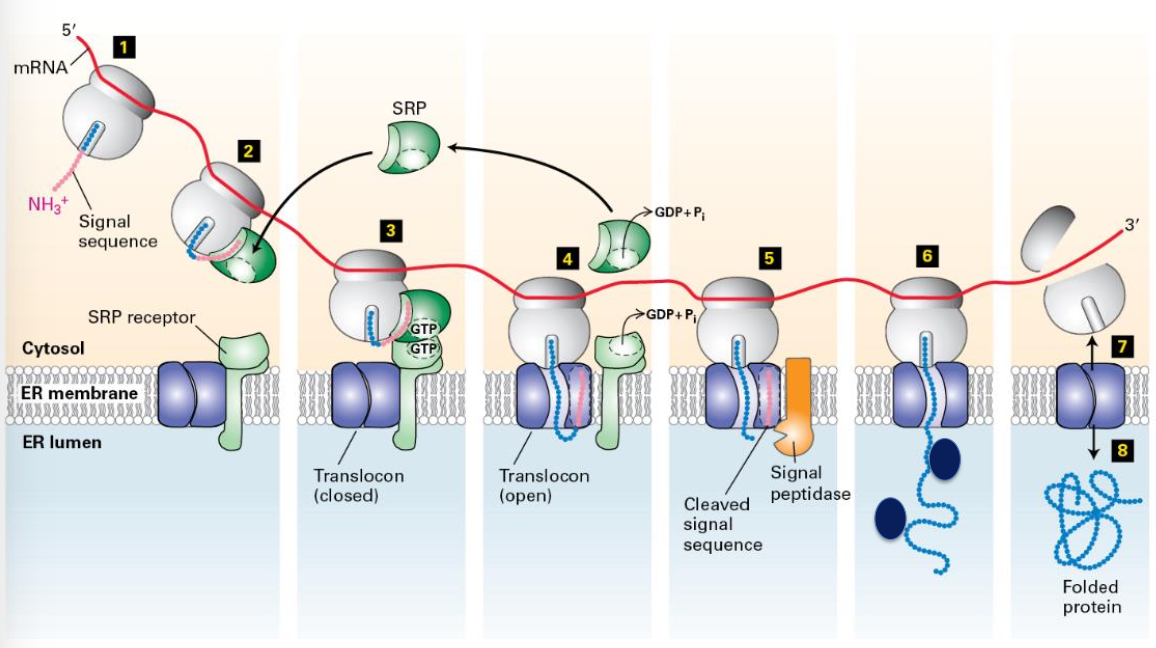
Cotranslational Translocation
Proteins are incorporated into microsomes during translation (cotranslational translocation). Transport of most secretory proteins into the ER lumen begins while the nascent protein is still bound to the ribosome.
Synthesis of Secretory Proteins on rough ER
mRNA binds to free ribosomes. Translation begins and the signal sequence is created.
continuous stretch of 6-12 amino acids at N terminus.
one of more positively charged amino acids next to this hydrophobic core
this hydrophobic core of the signal sequence is critical for interacting with the machinery that targets the protein to the ER membrane
Signal sequence recognized by a signal recognition particle and translation is temporarily arrested.
SRP binds SRP receptor (associated with translocon pore) .
Ribosome interacts with translocon. SRP dissociates from ribosome and receptor. Protein synthesis resumes.
Polypeptide translocates through the pore and enters the ER lumen. Signal sequence is cleaved by signal peptidase.
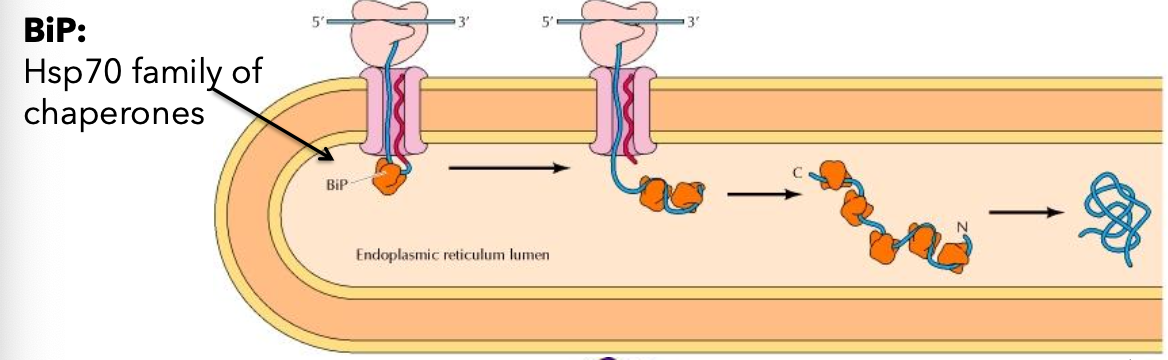
Signal sequence is cleaved by signal peptidase and is bound by BiP (chaperone protein) after translocation.
Functions of BiP
Dual role in protein processing; acts upon binding and release of ADP/ATP.
guards translocon pore when ADP bound
ADP bound: high substrate affinity
ATP bound: low substrate affinity
Integral Membrane Protein Synthesis
Orientation of integral membrane proteins determined during synthesis, influencing plasma membrane orientation.
the end that faces or is inside the ER lumen will face the extracellular space
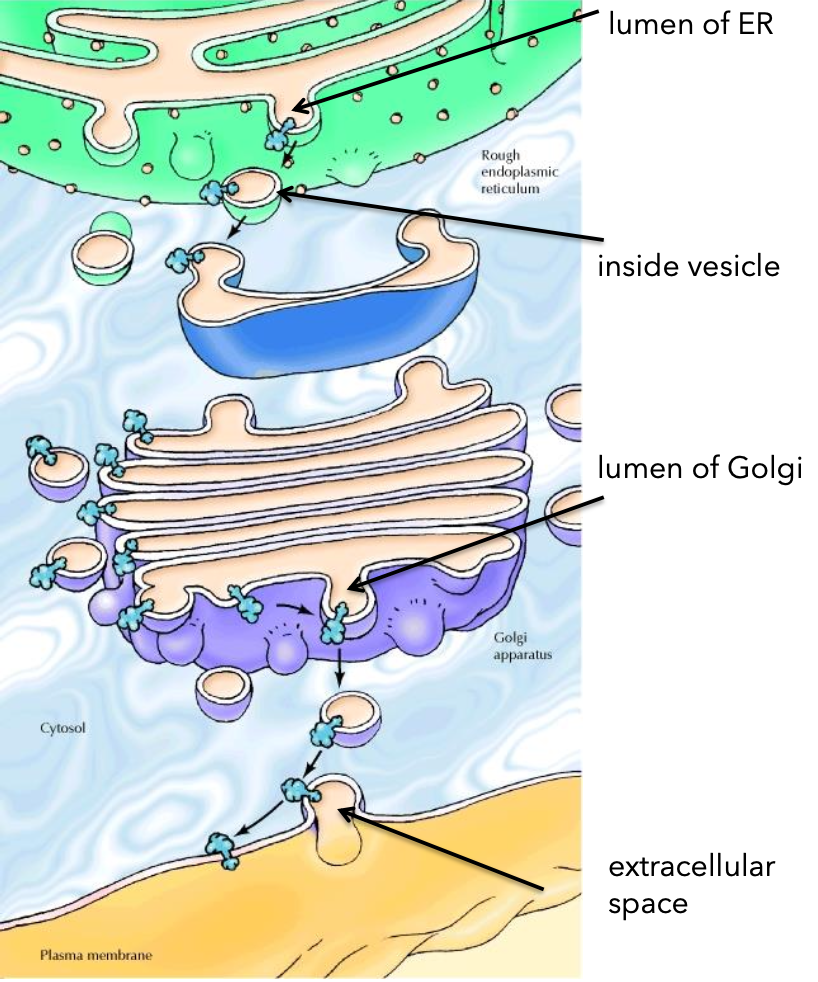
Topological Equivalence
Orientation concept: lumen of ER = Golgi lumen = extracellular space.
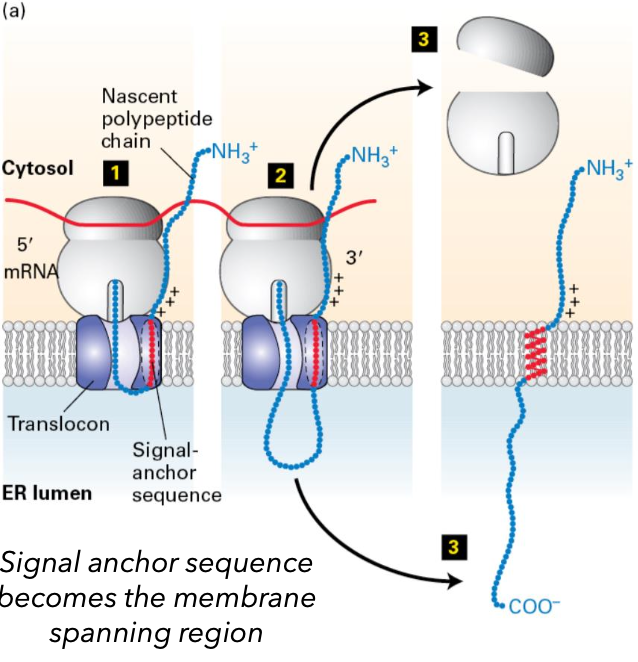
Insertion Mechanism of Membrane Proteins
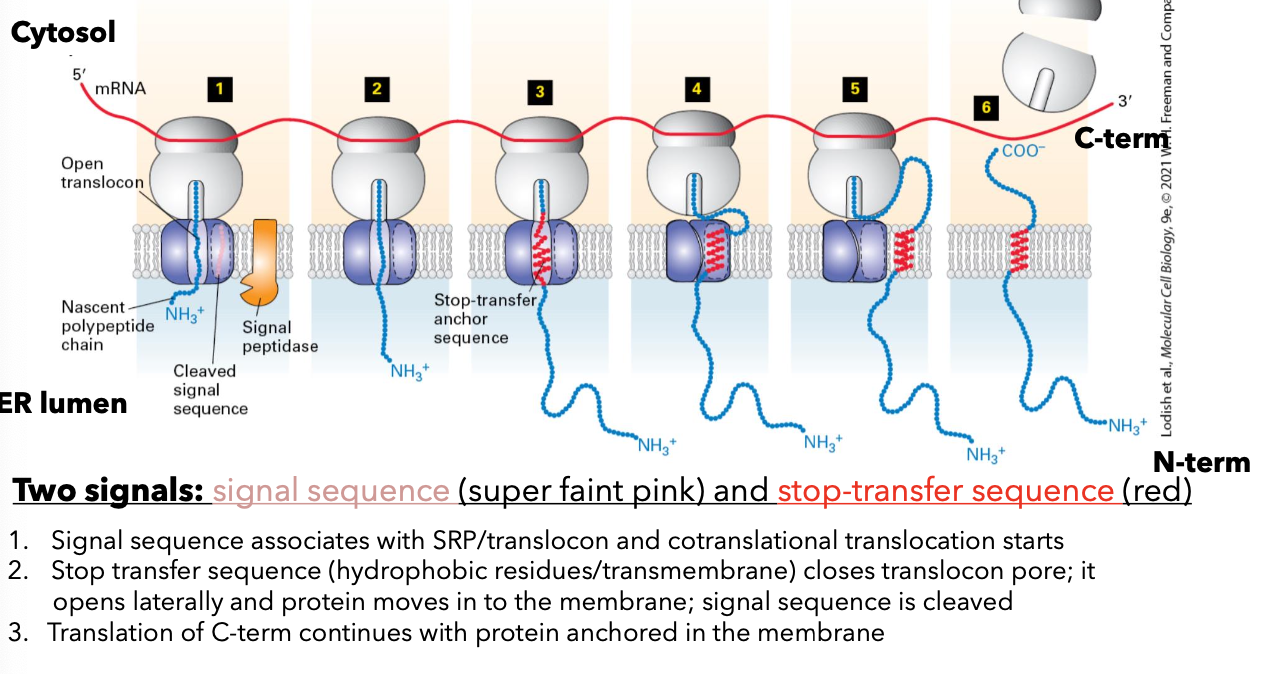
Signal sequences (an internal signal anchor sequence) is in the middle of the protein or at the COO- end (not at the NH3 end)
signal sequence targets the ribosome and peptide to the ER
Rjbsosome is transferred to open translocon.
NH3 end is in the cytosol
signal anchor sequence sticks in translocon
peptide elongated into ER lumen; carbohydrate is added
ribosome dissociated from the translocon; protein diffuses out of the translocon
Insertion of Multipass Proteins
Membrane spanning regions pass from the translocon into the membrance cotranslationally in order they energy from the ribosome
orientation of the first transmembrane segment is first established
initial engagement with the translocon in SRP/SRP receptor dependent manner
subsequent transmembrane segment assumes the opposite orientation
independent of SRP
Processing of Newly Synthesized Proteins
Specific Proteolytic cleavages in the ER, golgi complex, and secretory vesicles for protein maturation.
signal peptidase: cleaves signal sequence
Formation of Disulfide bond for stability and folding in the ER.
covalent bonds formed by the oxidative linkage of sulfhydryl groups, on two cysteine residues in the same or different polypeptide chains
functions: protein folding, increases stability of native structure
protein disulfide isomerase: promotes proper disulfide bond formation
Glycosylation: covalent addition of carbohydrates
Covalent addition of carbohydrates produces glycoproteins, facilitating structural stability and interactions.
functions incldue proper folding of proteins, structural stability, and produces an array of chemically distinct molecules at the cell surafce that are the basis of specific molecular interactions used in cell to cell adhesion and communication
Proper folding of polypeptide chains and assembly of multi-subunit proteins in the ER
Chaperone proteins like BiP play crucial roles in promoting protein folding and multi-subunit assembly.
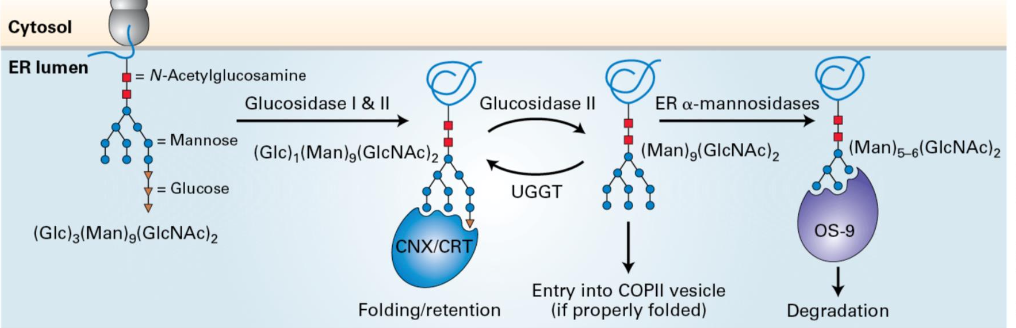
Quality Control in the ER
Modification of glycoproteins involves the removal of glucose by glucosidase II. Retain only a single glucose.
Glucose recognized and bound by ER chaperone calnexin.
Glucosidase II removes remaining glucose
Conformation sensing enzyme UGGT to determine if properly folded.
Unfolded proteins are targeted for degradation if improperly folded.
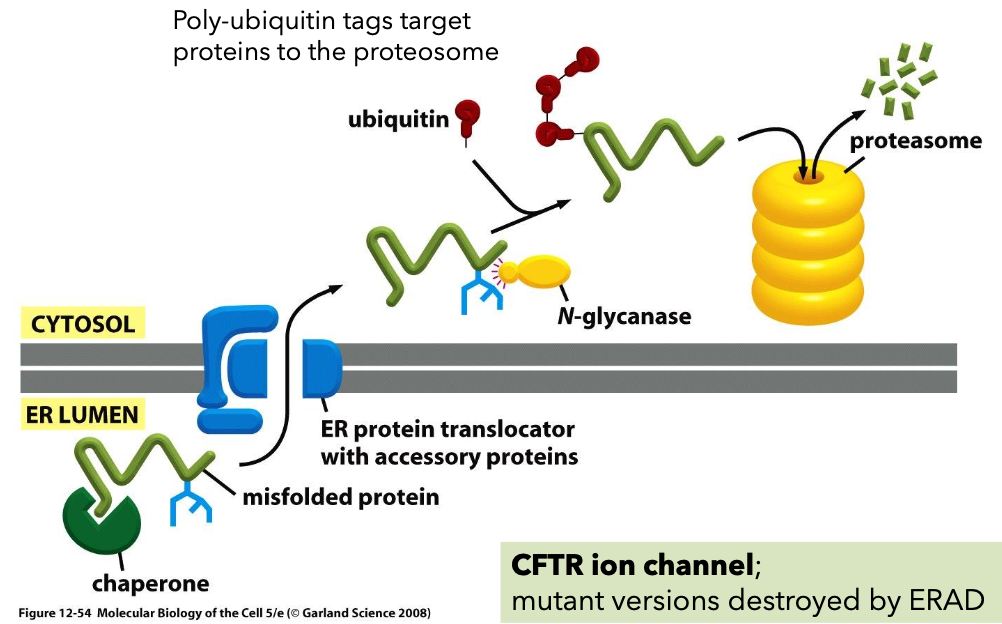
ER associated degradation Mechanisms
ER-associated degradation (ERAD) with poly-ubiquitin tags targeting proteins to the proteasome for disposal.
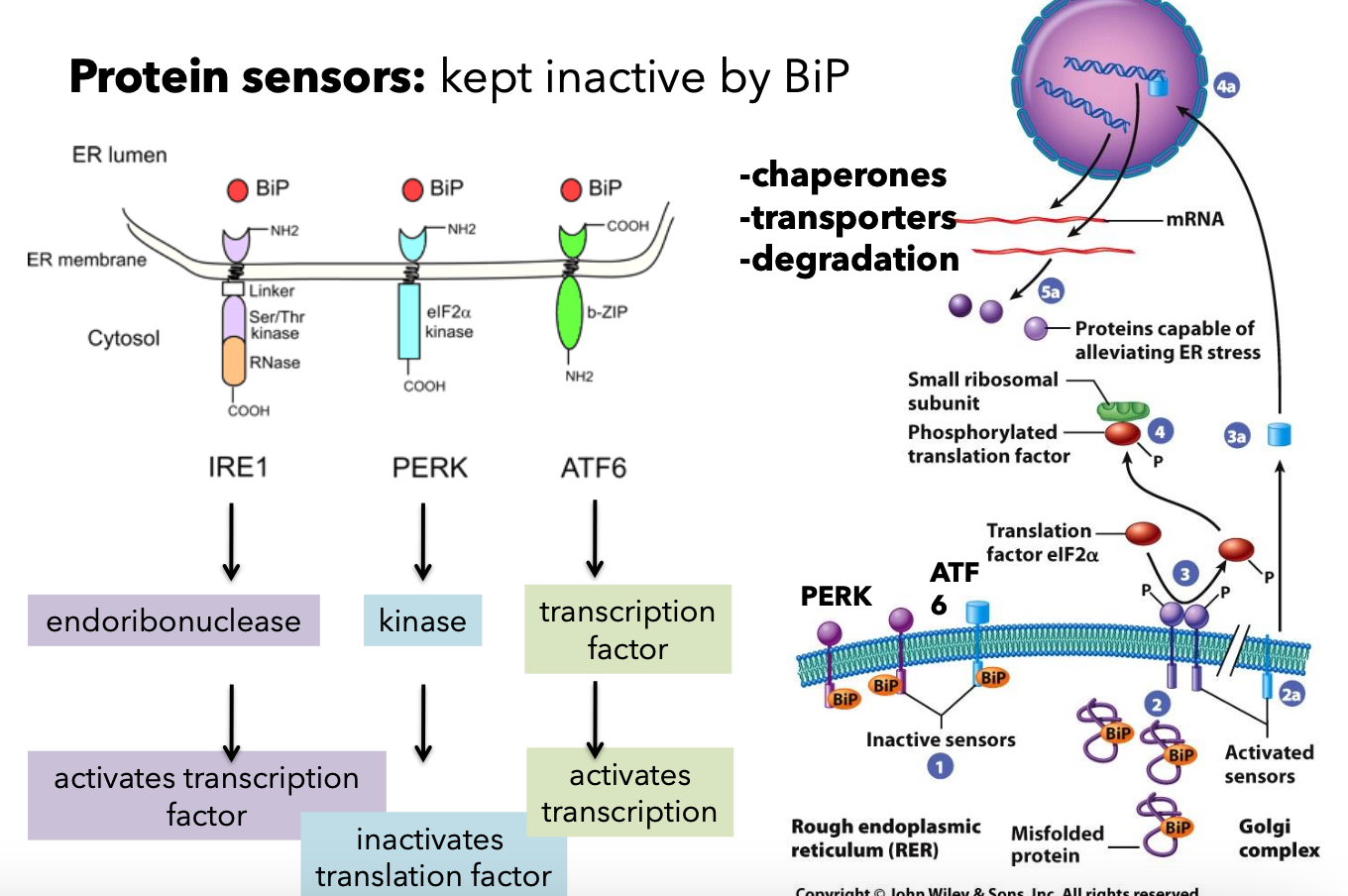
Unfolded Protein Response (UPR)
Activation of transcription factors in response to unfolded proteins, modulating chaperones and degradation pathways.