8. Omics techniques for target identification and development of biologics
Lecture overview
• How omics can be used for target discovery
• Omics techniques (genomics, transcriptomics, proteomics and metabolomics)
• Analysis of omics data for identification of targets
• Omics for personalised medicine
• Proteomics: focus and example applications
• How omics can help in discovery of relevant targets and develop biologics
• Conceptual understanding of some omics techniques
• Omics and some concepts of personalised medicine
Omics - Studies of the entire collection of a type of molecules. Omics means “a study of the totality of something”. The main four ones are genomics, transcriptomics, proteomics and metabolomics.
gene + -omics → genomics
protein + -omics → proteomics
metabolism + -omics → metabolomics
glyco- + -omics → glycomics
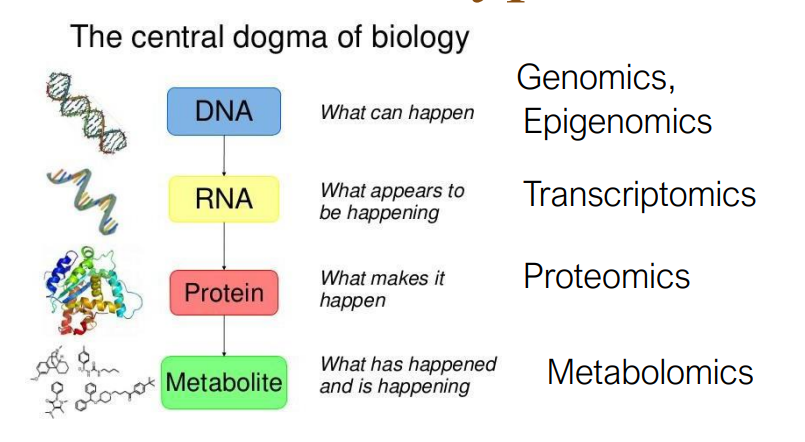
Genomics- The study of the set of genes contained in the chromosomes
Transcriptomics- The study of the set of mRNA molecules being expressed at a given time under specified conditions
Proteomics- The study of the set of proteins being expressed at a given time under specified conditions and their state of modification
Metabolomics- The study of the set of small molecules at a given time under specified conditions
Omics for target identification (target selection)
• Find molecular differences between healthy and disease
• Samples: Patient biopsy samples (from tissue which is quite invasive or liquid biopsy, blood sample), patient-derived cell lines (collect cells from a tumour), etc.
• Compare groups of samples to distinguish variation due to disease from normal population variation
• Find out which differences are causative to pinpoint suitable targets
Target discovery followed by biologics development
• Interaction partners of biopharmaceutical.
• Effect of biologics can be followed using omics
• Omics also useful for finding systemic side-effects.
Some techniques for Omics
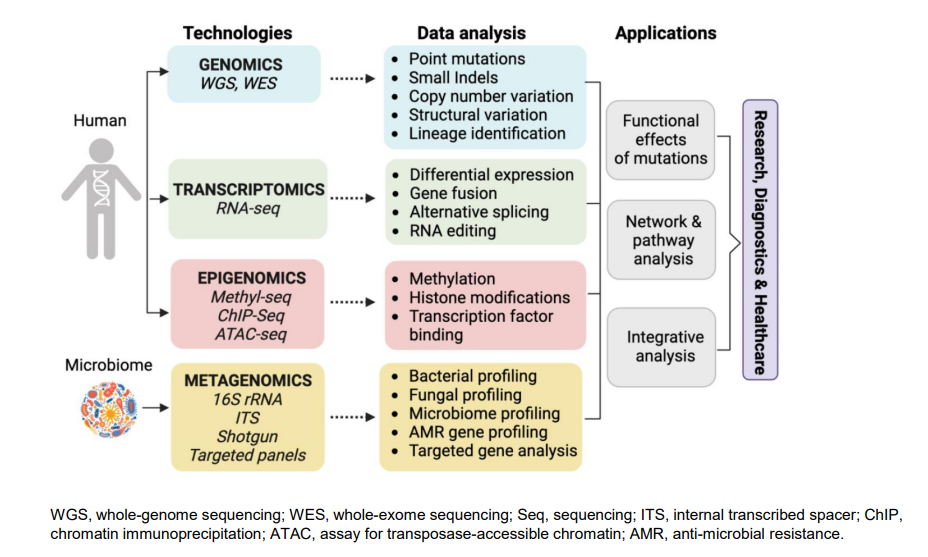
Genomics
- Whole genome Sequencing (WGS) and targeted resequencing using Next Generation Sequencing (NGS) (only target different regions of genome)
- Chip based variant detection and analyses using oligoneclotide probes (look for a small nucleic difference)
- Exome sequencing (NGS)
Transcriptomics
- RNA-sequencing (NGS)
- cDNA microarrays
- Single cell sequencing (NGS)
Proteomics
- Mass spectrometry (main method)
- Affinity-based proteomics
Metabolomics
- NMR
- Mass Spectrometry
Metagenomics
- Enviromental samples (NGS)
Single-cell omics
Epigenetics
- NGS
- Microarrays
Spatial omics
- Imaging
- NGS
NGS applications in human health
Genomics
- Find genome-level deviations that may be causing disease
- can be point mutations, indels, cross-overs, copy number variations in cancers
- Associations between genotype and phenotype
- GWAS (Genome-wide association studies)
- Large patient cohorts needed to obtain statistical power
Transcriptomics
- Look at the genes that are actually expressed under a certain condition
Transcriptomics technologies- The techniques used to study an organism’s whole
transcriptome, the sum of all of its RNA transcripts.
- The information content of an organism is recorded in the DNA of its genome and expressed through transcription.
- A transcriptome captures a snapshot in time of the total transcripts present in a cell.
Transcriptomics techniques include:
- DNA microarrays
- NGS technologies called RNA-Seq
- Transcription can also be studied at the level of individual cells by single-cell transcriptomics (ScRNASeq).
- Spatial transcriptomics where location of expression is captured by imaging together with transcript read out using probes or NGS
Microarrays and RNASeq are most common. Different single-cell techniques have gained increasing popularity.
RNA-sequencing vs microarrays
RNASeq: Involves the conversion of RNA into complementary DNA (cDNA), followed by sequencing the cDNA fragments using NGS technologies. Fragmentation step that are aligned. The sequence is built up.
Microarrays: create cDNA from RNA sample. Probes for the genes present on the microarray bind to a particular transcript. Quantitative analysis is carried out by the fluorescent signal.
Differential expression analyses – find differences between disease and control or subgroups of disese
- Heatmapping is used to visually represent patterns and relationships in large datasets
- To find differences between disease and control or subgroups of disease
- To find out if a gene is differentially expressed when comparing samples from different conditions
- Gene is found in higher or lower abundance
- Fluorescence in microarrays
- Read count in sequencing
Gene expression varies across tissues and conditions
- Have to consider where the gene is expressed
- Some genes are expressed in all tissues, while others only expressed in specific tissues/conditions.
- Tissue specific genes might be better targets to avoid secondary effects.
Network Analyses and Systems Biology: How genes work together?
- Genes/proteins do not work alone
- Often many genes/proteins change expression levels between conditions
- Networks (representation of complex systems (cells, tissues and organisms))and pathways enable us to interpret global patterns in the data (map all these findings into networks and pathways)
Gene set enrichment analyses (GSEA)- also called functional enrichment analysis or pathway enrichment analysis. Used to interpret gene expression data.
- Look at pathways or collection of genes that are connected to a pathway, disease etc.
- Look at the differentially expressed genes and see how they map onto the pathways.
- Look for differences that are likely not random → likely associated with the observed changes.
Things to consider for data analysis
- Sample size, needs to be large enough to get accurate data
- Batch effects/randomisation
- Correlation is not causation → need validation
- Bioinformatics
- Are the results reproducible and possible to replicate with other samples?
Proteomics- Closer to the phenotype
Isn’t RNA sequencing enough?
- Protein translation modifications (PTMs) affect protein function for example phosphorylation
- Protein localisation (secreted / membrane etc), different location might have functions
- Protein-protein interactions, important for protein function
- Sometimes low correlation between protein and RNA levels
How to measure protein levels?
- Mass spectrometry – based analysis (main method)
- Different types of Affinity reagents
- Antibodies (with different readouts, including MS)
What do to with quantitative proteomics data
• Similar to transcriptomics data (although RNA seq and MS proteomics data have different distributions)
• Differential abundance comparisons between sample groups
• Mapping to pathways etc
• Complicated by:
– Mapping peptides back to proteins and further to genes may be ambiguous
– Post-translational modifications, complicate analysis
Benefits of proteomics
• Possible to study relevant proteome for drug purposes, for example:
– Secreted proteins (secretome), be important for signilling or target
– Cell surface proteins
– Phosphoproteome for signalling
Pinpoint which part of the protein is important
Omics and precision medicine
- Some diseases are heterogeneous in the way that they have different mechanisms (even in the same tissue, e.g. cancer) etc.
- People are different. Some respond to treatment, while others don’t
→ Use omics approaches to stratify disease and patients
- Can we find relevant biomarkers to measure in the clinic? Develop ’Companion diagnostics’ to identify responders, patient that risk side effects and/or to follow treatment
- Find relevant biomarkers to distinguish from different forms of the disease, to find out which patients that respond/don’t respond to treatment etc.
→ Enable precision medicine (personalised medicine) so each patient can get the best possible treatment
Subclassification of disease using expression data
• Cancers are heterogenous. Subgroups may respond very differently to treatment (very different mechanism)
• Find markers for classification and personalised treatment
• Clustering of cancers using gene expression data may provide new subgroups (genetic typing can be done for known driver mutations in some cancers, but multiple mutations may have similar phenotypic effects), that can get different treatments
Metabolomics
• Reflecting the phenotype
• Analytically challenging to capture all metabolites
• Mass spectrometry and NMR main techniques
Summary
• Omics techniques can be used to find actionable molecular differences between sick and healthy and also further subtypes
• Data analysis critical
• Individual variation and co-variates need to be considered
• Complex network of biomolecules
• Choice of omics technique depends on nature of disease as well as availablility of samples. Rapid technology developments.
• Large multi-omics studies may pave the way for successful precision medicine