🍩AP BIO Chapter 20 - Phylogeny
Overview
20.1 Phylogenies show evolutionary relationships
20.2 Phylogenies are inferred from morphological and molecular data
20.3 Shared characters are used to construct phylogenetic trees
20.4 Molecular clocks help track evolutionary time
20.5 New information continues to revise our understanding of evolutionary history
BIG IDEAS: Phylogenetic trees and cladograms are models of common ancestry and evolutionary history that can be used to establish the relatedness of different species through analysis of similarities in morphology (Big Idea 1) or comparison of DNA and proteins (Big Idea 3)
We can decide in which category to place a species by comparing its traits with those of ___
- potential close relatives
]]Phylogeny:]]
The evolutionary history of a species or group of related species
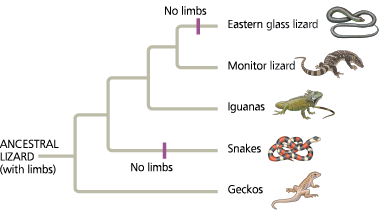
Convergent evolution of limbless bodies - A phylogeny based on DNA sequence data reveals that a legless body form evolved independently from legged ancestors in the lineages leading to the eastern glass lizard and to snakes
]]Systematics:]]
A scientific discipline focused on classifying organisms and determining their evolutionary relationships
==20.1 - Phylogenies show evolutionary relationships==
Linnaeus’s binomial classification system gives organisms two-part names: a genus plus a specific epithet
In the Linnaean system, species are grouped in increasingly broad taxa: Related genera are placed in the same family, families in orders, orders in classes, classes in phyla, phyla in kingdoms, and (more recently) kingdoms in domains
Systematists depict evolutionary relationships as branching phylogenetic trees. Many systematists propose that classification be based entirely on evolutionary relationships
Unless branch lengths are proportional to time or genetic change, a phylogenetic tree indicates only patterns of descent
Much information can be learned about a sTecies from its evolutionary history; hence, phylogenies are useful in a wide range of applications
]]Taxonomy:]]
A scientific discipline concerned with naming and classifying the diverse forms of life
]]Binomial:]]
A common term for the two-part, latinized format for naming a species, consisting of the genus and specific epithet; also called a binomen
]]Genus:]]
A taxonomic category above the species level, designated by the first word of a species’ two-part scientific name
]]Specific Epithet:]]
The second part of a binomial unique for each species within the genus
Species that appear to be closely related are grouped into the same ___
- genus
]]Family:]]
In Linnaean classification, the taxonomic category above genus
]]Orders:]]
In Linnaean classification, the taxonomic category above family
]]Class:]]
In Linnaean classification, the taxonomic category above order
]]Phyla:]]
In Linnaean classification, the taxonomic category above class
]]Kingdom:]]
In Linnaean classification, the taxonomic category above phyla
]]Domain:]]
A taxonomic category above the kingdom level; consists of archaea, bacteria, and eukarya
What are the levels in the Linnaean system? (from lowest to highest)
Genus, family, order, class, phylum, kingdoms, and domains
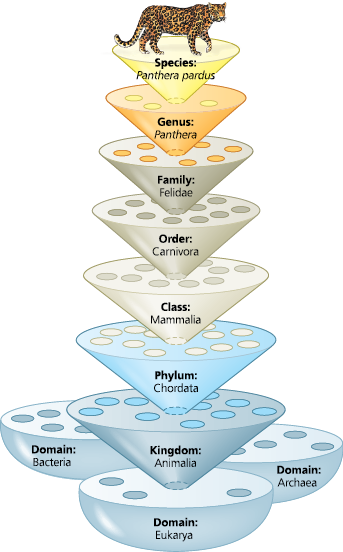
Linnaean classification - Panthera pardus
]]Taxon:]]
A named taxonomic unit at any given level of classification
]]Phylogenetic Tree:]]
A branching diagram that represents a hypothesis about the evolutionary history of a group of organisms
The connection between classification and phylogeny - Hierarchical classification can reflect the branching patterns of phylogenetic trees. This tree traces possible evolutionary relationships between some of the taxa within order Carnivora, itself a branch of class Mammalia. The branch point 1 represents the most recent common ancestor of all members of the weasel (Mustelidae) and dog (Canidae) families. The branch point 2 represents the most recent common ancestor of coyotes and gray wolves
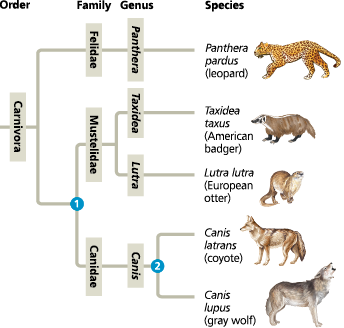
==What does this phylogenetic tree indicate about the evolutionary relationships between the leopard, badger, and wolf?==
The branching pattern of the tree indicates that the badger and the wolf share a common ancestor that is more recent than the ancestor these two animals share with the leopard
]]Branch Points:]]
The representation on a phylogenetic tree of the divergence of two or more taxa from a common ancestor. A branch point is usually shown as a dichotomy in which a branch representing the ancestral lineage splits (at the branch point) into two branches, one for each of the two descendant lineages
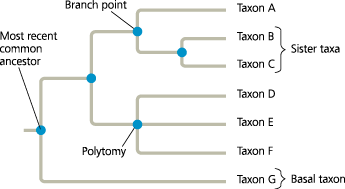
How to read a phylogenetic tree
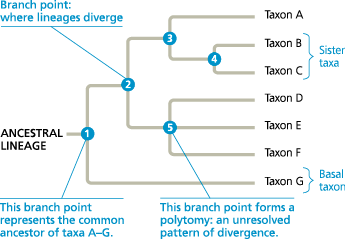
Redraw this tree, rotating the branches around branch points 2 and 4. Does your new version tell a different story about the evolutionary relationships between the taxa? Explain
The new version (shown below) does not alter any of the evolutionary relationships shown in Figure 20.5. For example, B and C remain sister taxa, taxon A is still as closely related to taxon B as it is to taxon C, and so on 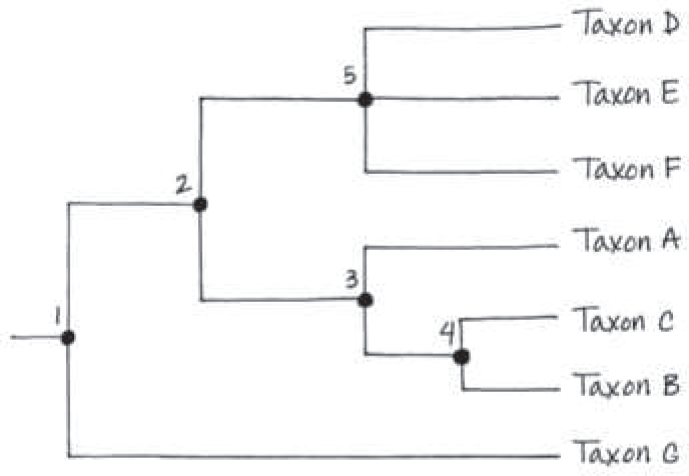
]]Sister Taxa:]]
Groups of organisms that share an immediate common ancestor and hence are each other’s closest relatives
]]Rooted:]]
Describing a phylogenetic tree that contains a branch point (often, the one farthest to the left) representing the most recent common ancestor of all taxa in the tree
]]Basal Taxon:]]
In a specified group of organisms, a taxon whose evolutionary lineage diverged early in the history of the group
]]Polytomy:]]
In a phylogenetic tree, a branch point from which more than two descendant taxa emerge. A polytomy indicates that the evolutionary relationships between the descendant taxa are not yet clear
What are three key points about phylogenetic trees?
They are intended to show patterns of descent, not phenotypic similarity, the sequence of branching in a tree does not necessarily indicate the actual (absolute) ages of the particular species, and we should not assume that a taxon on a phylogenetic tree evolved from the taxon next to it
C. S. Baker and S. R. Palumbi purchased 13 samples of “whale meat” from Japanese fish markets. They sequenced part of the mitochondrial DNA (mtDNA) from each sample and compared their results with the comparable mtDNA sequence from known whale species. To infer the species identity of each sample, the team constructed a gene tree, a phylogenetic tree that shows patterns of relatedness among DNA sequences rather than among taxa
Results: 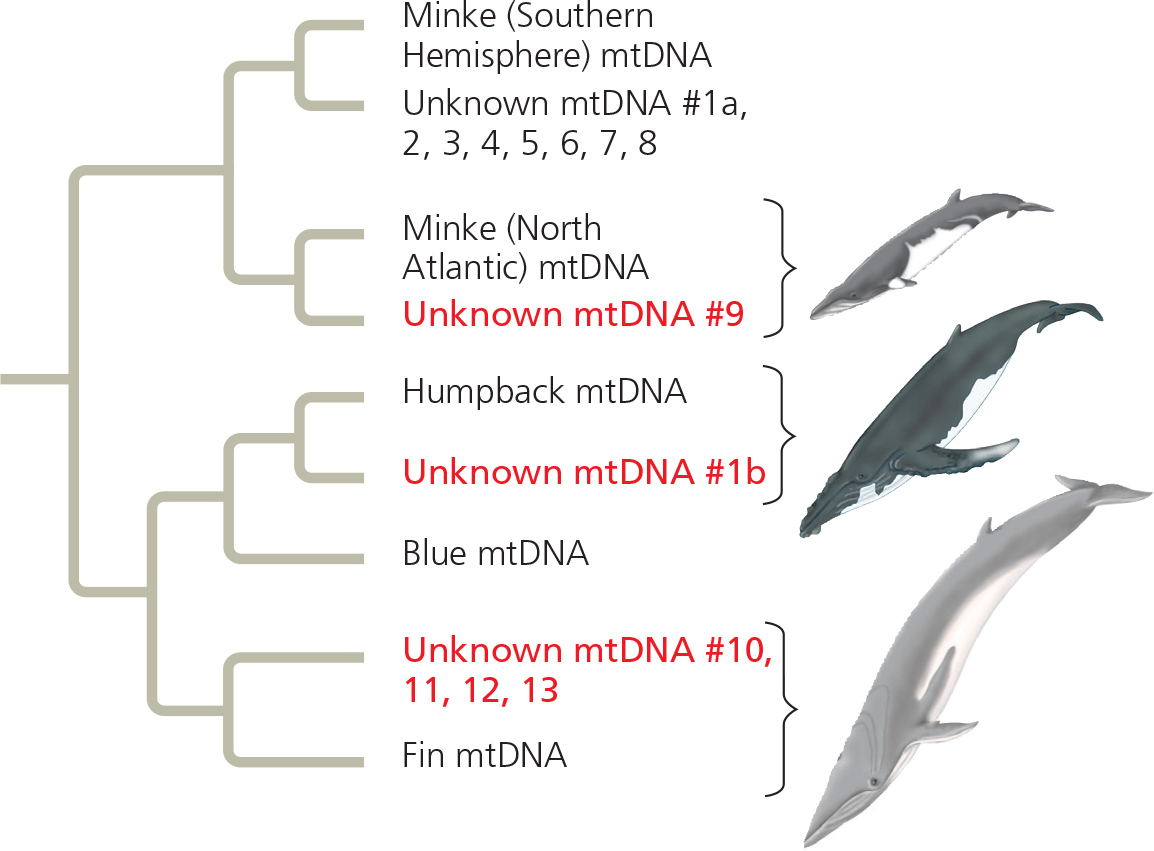 The mtDNA sequences of six of the unknown samples (in red) were most similar to mtDNA sequences of whales that are not legal to harvest, indicating that the unknown samples were from illegally harvested whales
The mtDNA sequences of six of the unknown samples (in red) were most similar to mtDNA sequences of whales that are not legal to harvest, indicating that the unknown samples were from illegally harvested whales
==Which levels of the classification in Figure 20.3 do humans share with leopards?==
We are classified the same from the domain level to the class level; both the leopard and human are mammals. Leopards belong to order Carnivora, whereas humans do not
==Which of the trees shown here depicts an evolutionary history different from the other two?== 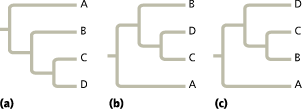
The tree in (c) shows a different pattern of evolutionary relationships. In (c), C and B are sister taxa, whereas C and D are sister taxa in (a) and (b)
==Suppose new evidence indicates that taxon E in Figure 20.5 is the sister taxon of a group consisting of taxa D and F. Redraw the tree accordingly==
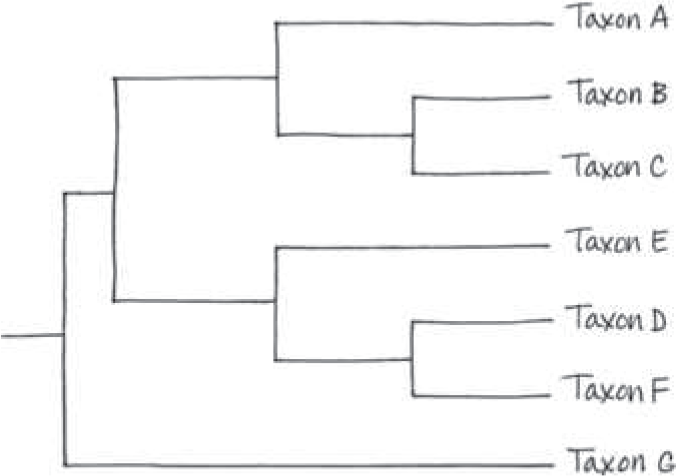
==Humans and chimpanzees are sister species. Explain what that means==
The fact that humans and chimpanzees are sister species indicates that we share a more recent common ancestor with chimpanzees than we do with any other living primate species. But that does not mean that humans evolved from chimpanzees, or vice versa; instead, it indicates that both humans and chimpanzees are descendants of that common ancestor
==20.2 - Phylogenies are inferred from morphological and molecular data==
Organisms with similar morphologies or DNA sequences are likely to be more closely related than organisms with very different structures and genetic sequences
To infer phylogeny, homology (similarity due to shared ancestry) must be distinguished from analogy (similarity due to convergent evolution)
Computer programs are used to align comparable DNA sequences and to distinguish molecular homologies from coincidental matches between taxa that diverged long ago
The morphological divergence between related species can be great while their genetic divergence is ___
- small
]]Analogy:]]
Similarity between two species that is due to convergent evolution rather than to descent from a common ancestor with the same trait
Convergent evolution occurs when similar environmental pressures and natural selection produce similar (analogous) ___ in organisms from different evolutionary lineages
- adaptations
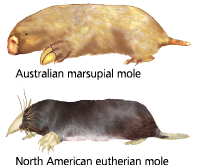
Convergent evolution in borrowers - A long body, large front paws, small eyes, and a pad of thick skin that protects the nose all evolved independently in these species
Compare bats with birds and cats
Bat wings are stretched membranes while bird wings have feathers. Bats are more related to cats, and a bat’s wing is more similar to the forelimbs of cats and other mammals than to a bird’s wing
With respect to flight, a bat’s wing is ___, not _, to a bird’s wing
- analogous; homologous
]]Homoplasy:]]
A similar (analogous) structure or molecular sequence that has evolved independently in two species
The more elements that are similar in two complex structures, the ___ likely it is that the structures evolved from a common ancestor
- more
If genes in two organisms share many portions of their nucleotide sequences, it is likely that the genes are ___
- homologous
Aligning segments of DNA - Systematists search for similar sequences along DNA segments from two species (only one DNA strand is shown for each species). In this example, 11 of the original 12 bases have not changed since the species diverged. Hence, those portions still align once the length is adjusted
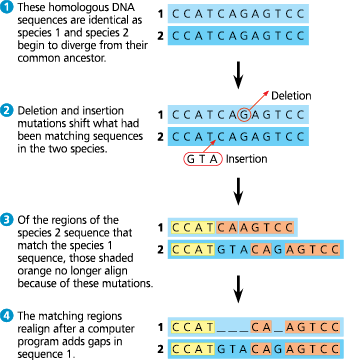
Why do distantly related species usually have different bases at many sites and have different lengths?
Insertions and deletions accumulate over long periods of time
How are the Australian mole and North American mole related?
They have morphological differences, but the high degree of gene sequence similarity indicates they are closely related
]]Molecular Homoplasy:]]
Organisms that do not appear to be closely related, the bases that their otherwise very different sequences happen to share may simply be coincidental matches
==Why might you expect organisms that are not closely related to nevertheless share roughly 25% of their bases?==
There are four possible bases (A, C, G, T) at each nucleotide position. If the base at each position depends on chance, not common descent, we would expect roughly one out of four (25%) of them to be the same
A molecular homoplasy - These two DNA sequences from organisms that are not closely related coincidentally share 23% of their bases. Statistical tools have been developed to determine whether DNA sequences that share more than 25% of their bases do so because they are homologous

==Decide whether each of the following pairs of structures more likely represents analogy or homology, and explain your reasoning: (a) a porcupine’s quills and a cactus’s spines; (b) a cat’s paw and a human’s hand; (c) an owl’s wing and a hornet’s wing==
(a) Analogy, since porcupines and cacti are not closely related and since most other animals and plants do not have similar structures; (b) homology, since cats and humans are both mammals and have homologous forelimbs, of which the hand and paw are the lower part; (c) analogy, since owls and hornets are not closely related and since the structure of their wings is very different
==Suppose that two species, A and B, have similar appearances but very divergent gene sequences, while species B and C have very different appearances but similar gene sequences. Which pair of species is more likely to be closely related: A and B, or B and C? Explain==
Species B and C are more likely to be closely related. Small genetic changes (as between species B and C) can produce divergent physical appearances, but if many genes have diverged greatly (as in species A and B), then the lineages have probably been separate for a long time
==Why is it necessary to distinguish homology from analogy to infer phylogeny?==
Homologous characters result from shared ancestry. As organisms diverge over time, some of their homologous characters will also diverge. The homologous characters of organisms that diverged long ago typically differ more than do the homologous characters of organisms that diverged more recently. As a result, differences in homologous characters can be used to infer phylogeny. In contrast, analogous characters result from convergent evolution, not shared ancestry, and hence can give misleading estimates of phylogeny
==20.3 - Shared characters are used to construct phylogenetic trees==
A clade is a monophyletic group that includes an ancestral species and all of its descendants
Clades can be distinguished by their shared derived characters
Branch lengths can be proportional to amount of evolutionary change or time
Among phylogenies, the most parsimonious tree is the one that requires the fewest evolutionary changes
Well-supported phylogenetic hypotheses are consistent with a wide range of data
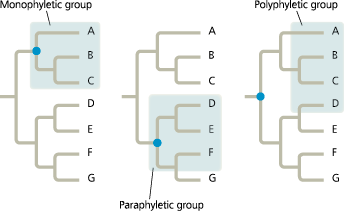
Do analogous or homologous features reflect evolutionary history?
Only homology reflects evolutionary history
]]Cladistics:]]
An approach to systematics in which organisms are placed into clades based primarily on common descent
]]Clades:]]
A group of species that includes an ancestral species and all of its descendants
]]Monophyletic:]]
Pertaining to a group of taxa that consists of a common ancestor and all of its descendants. A monophyletic taxon is equivalent to a clade
]]Paraphyletic:]]
Pertaining to a group of taxa that consists of a common ancestor and some, but not all, of its descendants
]]Polyphyletic:]]
Pertaining to a group of taxa that includes distantly related organisms but doe snot include their most recent common ancestor
Monophyletic, paraphyletic, and polyphyletic groups
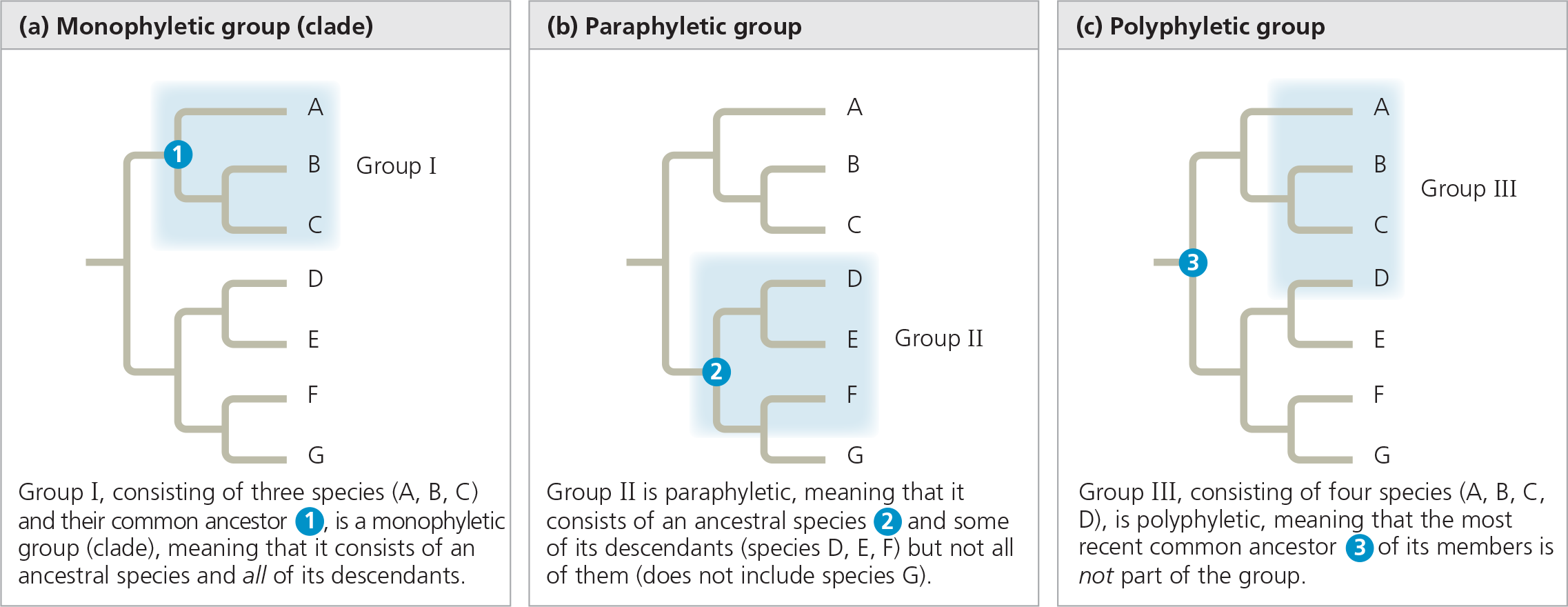
In a ___ group, the most recent common ancestor of all members of the group is part of the group, whereas in a _ group, the most recent common ancestor is not part of the group
- paraphyletic; polyphyletic
Paraphyletic vs. polyphyletic groups
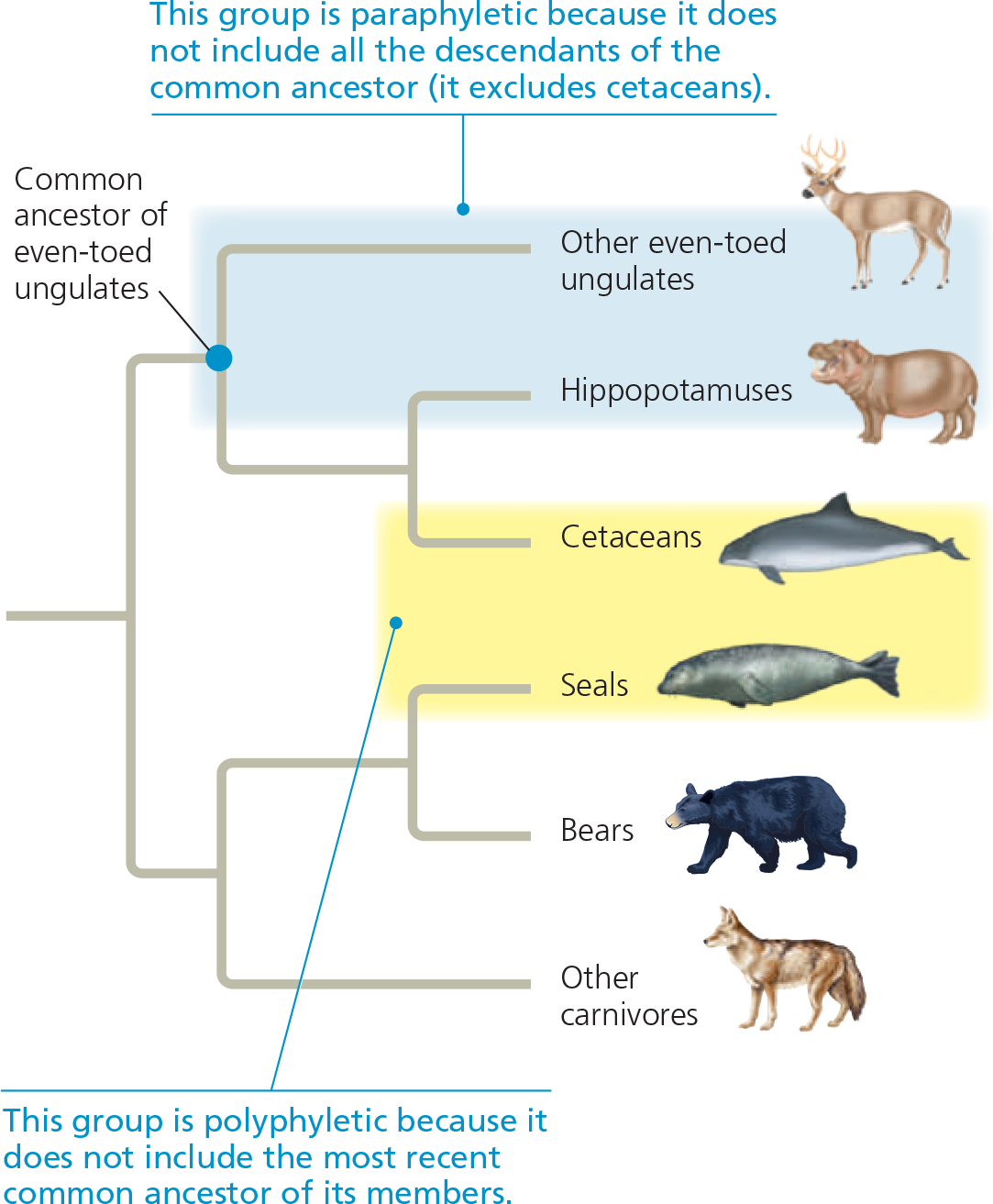
==Circle the branch point that represents the most recent common ancestor of cetaceans and seals. Explain why that ancestor would not be part of a cetacean-seal group defined by their similar body forms==
You should have circled the branch point that is drawn farthest to the left (the common ancestor of all taxa shown). Both cetaceans and seals descended from terrestrial lineages of mammals, indicating that the cetacean-seal common ancestor had legs and lacked a streamlined body form. As a result, that ancestor would not be part of the cetacean-seal group
]]Shared Ancestral Character:]]
A character that is shared by members of a particular clade but that originated in an ancestor that is not a member of that clade (backbone)
]]Shared Derived Character:]]
An evolutionary novelty that is unique to a particular clade; shared by organisms but not found in their ancestors (hair)
]]Outgroup:]]
A species or group of species from an evolutionary lineage that is known to have diverged before the lineage that contains the group of species being studied. An outgroup is selected so that its members are closely related to the group of species being studied, but not as closely related as any study-group members are to each other
]]Ingroup:]]
A species or group of species whose evolutionary relationships are being examined in a given analysis
Using derived characters to infer phylogeny - The derived characters used here include the amnion, a membrane that encloses the embryo inside a fluid-filled sac. Note that a different set of characters could lead us to infer a different phylogenetic tree
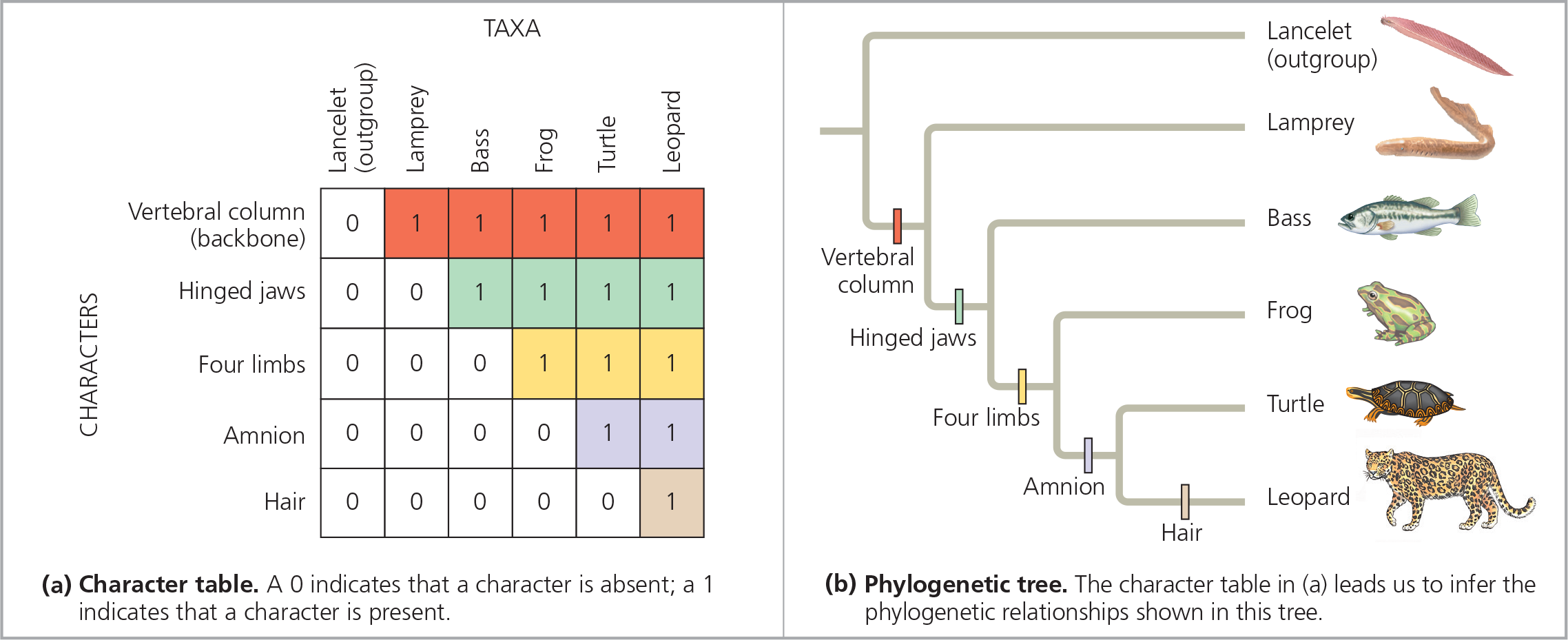
==In (b), circle the most inclusive clade for which a hinged jaw is a shared ancestral character==
You should have circled the frog, turtle, and leopard lineages, along with their most recent common ancestor
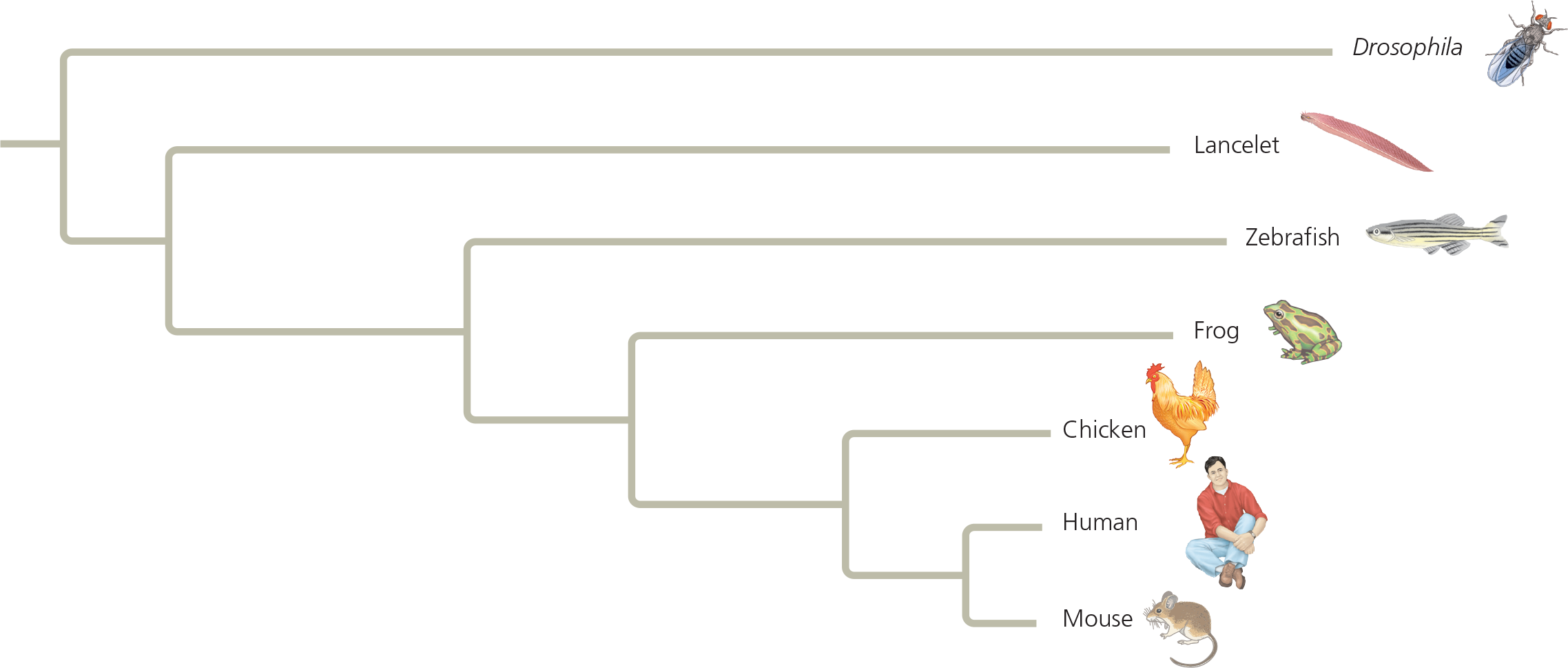
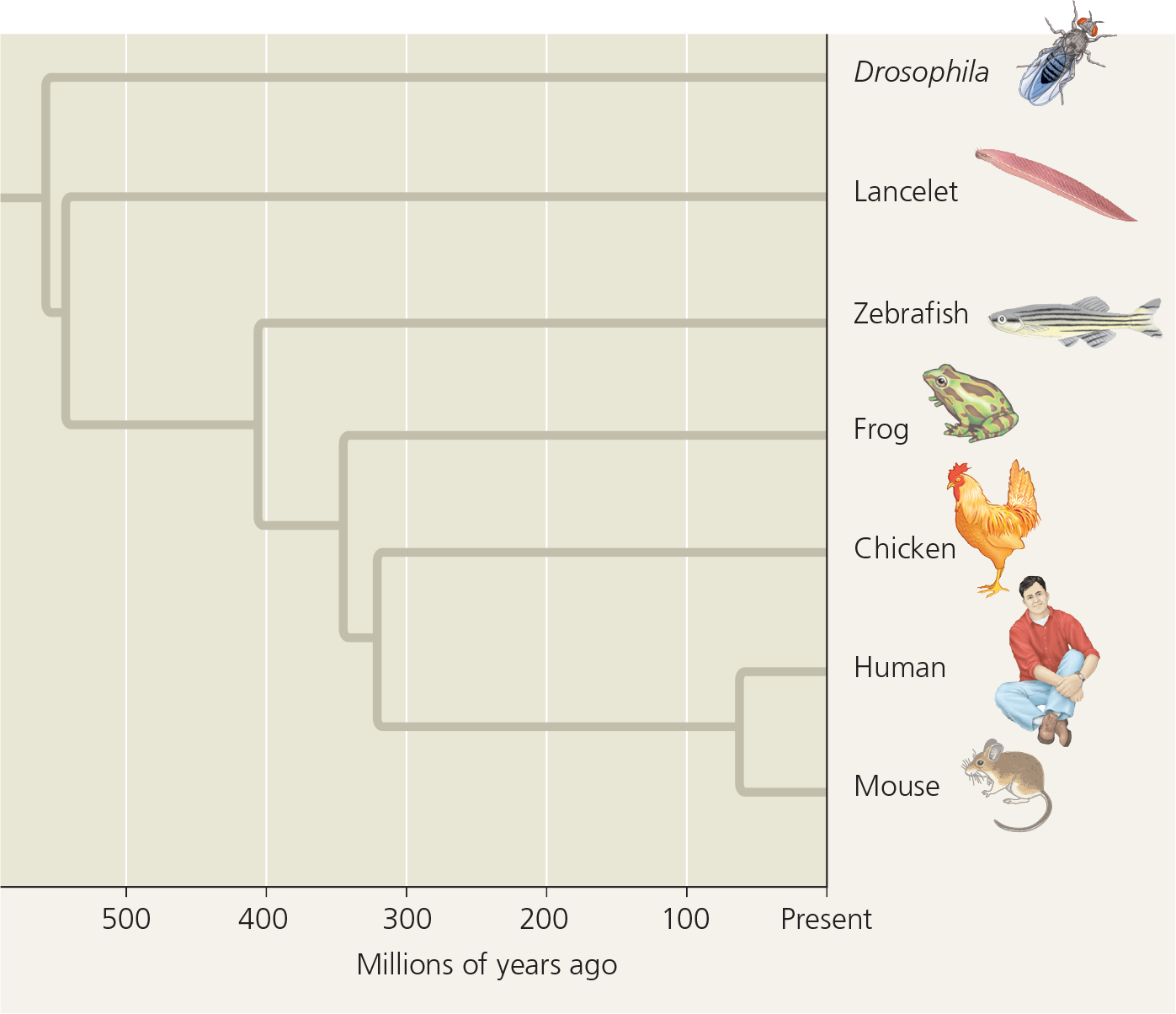
Branch lengths can represent genetic change - This tree was constructed by comparing sequences of homologs of a gene that plays a role in development; Drosophila was used as an outgroup. The branch lengths are proportional to the amount of genetic change in each lineage; varying branch lengths indicate that the gene has evolved at different rates in different lineages
Branch lengths can indicate time - The branch points are mapped to dates based on fossil evidence. Thus, the branch lengths are proportional to time. Each lineage has the same total length from the base of the tree to the branch tip, indicating that all the lineages have diverged from the common ancestor for equal amounts of time
]]Maximum Parsimony:]]
A principle that states when considering multiple explanations for an observation, one should first investigate the simplest explanation that is consistent with the facts
In the case of trees based on morphology, the most parsimonious tree requires the fewest ___, as measured by the origin of shared derived morphological characters. For phylogenies based on DNA, the most parsimonious tree requires the fewest _
- evolutionary events; base changes
Follow the numbered steps as we apply the principle of parsimony to a hypothetical phylogenetic problem involving three closely related bird species 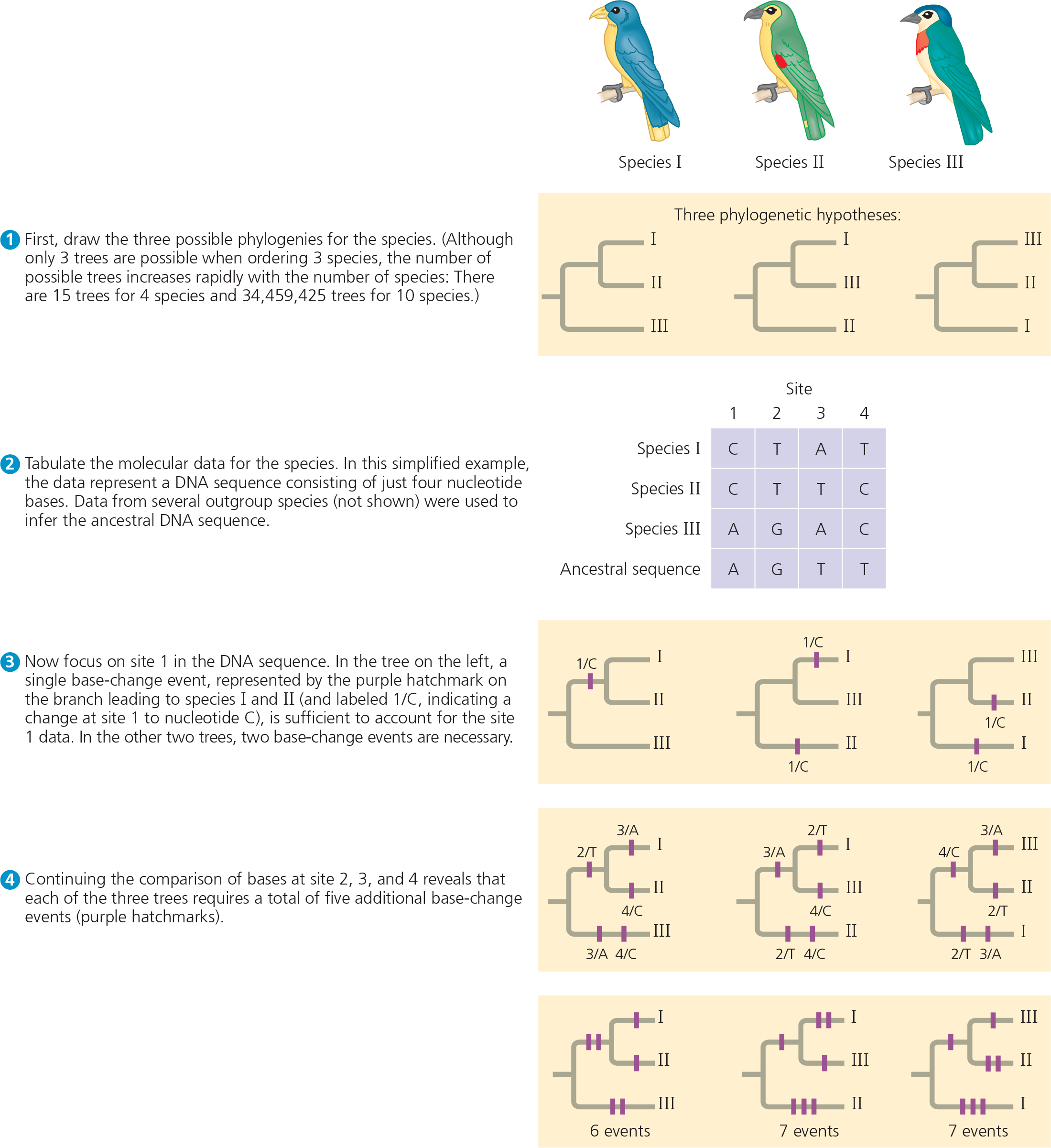
To identify the most parsimonious tree, we total all of the base-change events noted in steps 3 and 4. We conclude that the first tree is the most parsimonious of the three possible phylogenies. (In a real example, many more sites would be analyzed. Hence, the trees would often differ by more than one base-change event.)
Any phylogenetic tree represents a ___ about how the organisms in the tree are related to one another
- hypothesis
]]Phylogenetic Bracketing:]]
Inferencing the likelihood of unknown traits in organisms based on their position in a phylogenetic tree
A phylogenetic tree of birds and their close relatives
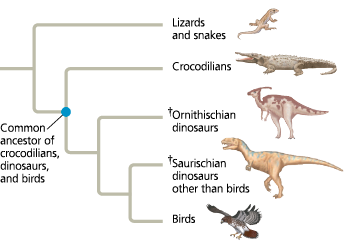 († indicates extinct lineages)
(† indicates extinct lineages)
==What is the most basal taxon represented in this tree?==
The lizard and snake lineage is the most basal taxon shown (closest to the root of the tree)
]]Brooding:]]
A behaviour in which a parent warms the eggs with its body
A crocodile guards its nest - After building its nest mound, this female African dwarf crocodile will care for the eggs until they hatch

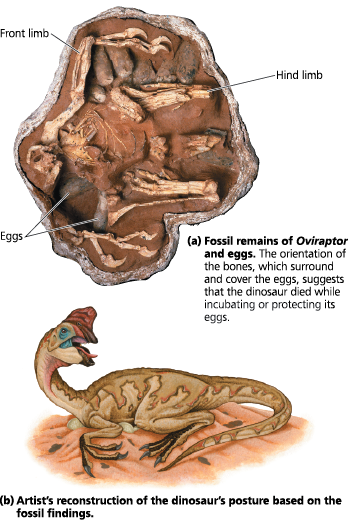
How have fossils supported the idea of brooding?
A fossil embryo of an Oviraptor dinosaur was found, still inside its egg. Researchers suggested that the Oviraptor dinosaur preserved in this second fossil died while incubating or protecting its eggs
Fossil support for a phylogenetic prediction: Dinosaurs built nests and brooded their eggs
What things rarely fossilize?
Soft tissues and internal organs such as the heart
==To distinguish a particular clade of mammals within the larger clade that corresponds to class Mammalia, would hair be a useful character? Why or why not?==
No; hair is a shared ancestral character common to all mammals and thus is not helpful in distinguishing different mammalian subgroups
==The most parsimonious tree of evolutionary relationships can be inaccurate. How can this occur?==
The principle of maximum parsimony states that the hypothesis about nature we investigate first should be the simplest explanation found to be consistent with the facts. Actual evolutionary relationships may differ from those inferred by parsimony owing to complicating factors such as convergent evolution
==Draw a phylogenetic tree that includes the relationships from Figure 20.16 and those shown here. Traditionally, all the taxa shown besides birds and mammals were classified as reptiles. Would a cladistic approach support that classification?== 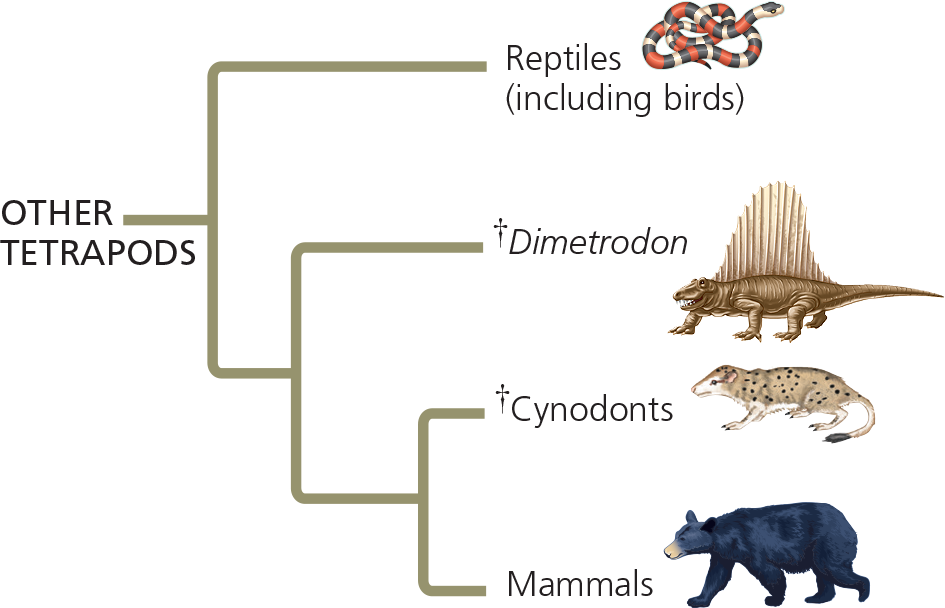
The traditional classification provides a poor match to evolutionary history, thus violating the basic principle of cladistics—that classification should be based on common descent. Both birds and mammals originated from groups traditionally designated as reptiles, making reptiles (as traditionally delineated) a paraphyletic group. These problems can be addressed by removing Dimetrodon and cynodonts from the reptiles and by regarding birds as a group of reptiles (specifically, as a group of dinosaurs) 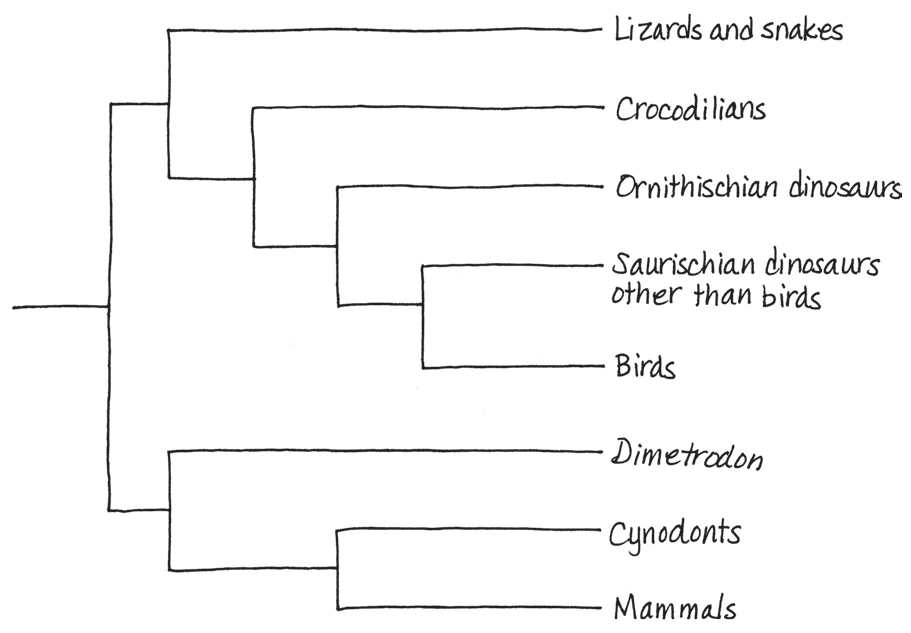
==Explain the logic of using shared derived characters to infer phylogeny==
All features of organisms arose at some point in the history of life. In the group in which a new feature first arose, that feature is a shared derived character that is unique to that clade. The group in which each shared derived character first appeared can be determined, and the resulting nested pattern can be used to infer the evolutionary history
==20.4 - Molecular clocks help track evolutionary time==
Some regions of DNA change at a rate consistent enough to serve as a molecular clock, in which the amount of genetic change is used to estimate the date of past evolutionary events. Other DNA regions change in a less predictable way
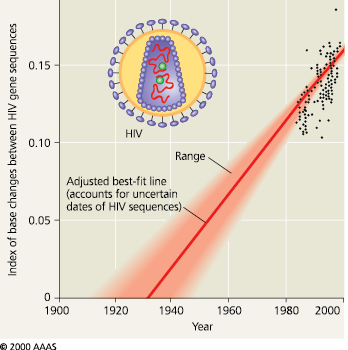
Molecular clock analyses suggest that the most common strain of HIV jumped from primates to humans in the early 1900s
]]Molecular Clock:]]
A method of estimating the time required for a given amount of evolutionary change, based on the observation that some regions of genomes evolve at constant rates
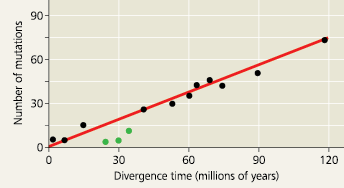
A molecular clock for mammals - The number of accumulated mutations in seven proteins has increased over time in a consistent manner for most mammal species. The three green data points represent primate species, whose proteins appear to have evolved more slowly than those of other mammals. The divergence time for each data point was based on fossil evidence
No gene marks time with complete precision, And even those genes that seem to act as reliable molecular clocks are accurate only in the statistical sense of showing a fairly smooth ___ rate of change. There may be _ over time from this
- average; deviations
The same gene may evolve at ___ rates in different groups of organisms
- different
When comparing genes that are clocklike, the rate of the clock may vary ___ from one gene to another
- greatly
What influences how genes grow quickly or slowly?
Gene growth depends on how important the amino acid sequence is to survival. If the exact sequence of amino acids that a gene specifies is essential to survival, most of the mutational changes will be harmful and only a few will be neutral. As a result, such genes change only slowly. But if the exact sequence of amino acids is less critical, fewer of the new mutations will be harmful and more will be neutral. Such genes change more quickly
Dating the origin of HIV-1 M - The black data points are based on DNA sequences of an HIV gene in patients’ blood samples. (The dates when these individual HIV gene sequences arose are not certain because a person can harbor the virus for years before symptoms occur.) Projecting the gene’s rate of change backward in time by this method suggests that the virus originated in the 1930s
==What is a molecular clock? What assumption underlies the use of a molecular clock?==
A molecular clock is a method of estimating the actual time of evolutionary events based on numbers of base changes in genes that are related by descent. It is based on the assumption that the regions of genomes being compared evolve at constant rates
==Explain how numerous base changes could occur in an organism’s DNA yet have no effect on its survival and reproduction==
There are many portions of the genome that do not code for genes; mutations that alter the sequence of bases in such regions could accumulate without affecting an organism’s survival and reproduction. Even in coding regions of the genome, some mutations may not have a critical effect on genes or proteins
==Suppose a molecular clock dates the divergence of two taxa at 80 million years ago, but new fossil evidence shows that the taxa diverged at least 120 million years ago. Explain how this could happen==
The gene (or genes) used for the molecular clock may have evolved more slowly in these two taxa than in the species used to calibrate the clock; as a result, the clock would underestimate the time at which the taxa diverged from each other
==Describe some assumptions and limitations of molecular clocks==
A key assumption of molecular clocks is that nucleotide substitutions occur at fixed rates, and hence the number of nucleotide differences between two DNA sequences is proportional to the time since the sequences diverged from each other. Some limitations of molecular clocks: No gene marks time with complete precision; natural selection can favor certain DNA changes over others; nucleotide substitution rates can change over long periods of time (causing molecular clock estimates of when events in the distant past occurred to be highly uncertain); and the same gene can evolve at different rates in different organisms
==20.5 - New information continues to revise our understanding of evolutionary history==
Past classification systems have given way to the current view of the tree of life, which consists of three great domains: Bacteria, Archaea, and Eukarya
Phylogenies based in part on rRNA genes suggest that eukaryotes are most closely related to archaea, while data from some other genes suggest a closer relationship to bacteria
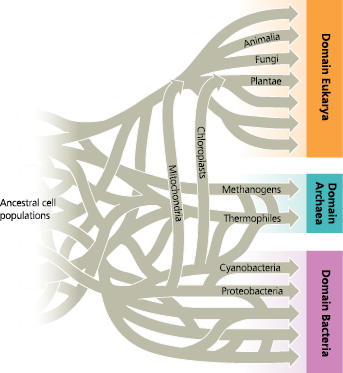
Genetic analyses indicate that extensive horizontal gene transfer has occurred throughout the evolutionary history of life
Identify the five kingdoms that were recognized on the late 1960s
Monera (prokaryotes), Protista (a diverse kingdom consisting mostly of unicellular organisms), Plantae, Fungi, and Animalia
Identify the three domains that are a taxonomic level higher than the kingdoms
Bacteria, Archaea, and Eukarya
The three domains of life - This phylogenetic tree is based on sequence data for rRNA and other genes. For simplicity, only some of the major branches in each domain are shown. Lineages within Eukarya that are dominated by multicellular organisms (plants, fungi, and animals) are in bold red type, while the two lineages denoted by an asterisk are based on DNA from cellular organelles. All other lineages consist solely or mainly of single-celled organisms
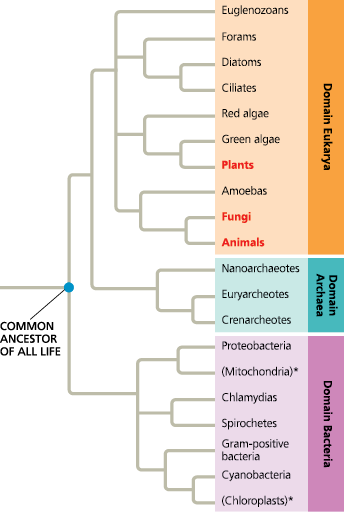
==After reviewing endosymbiont theory (see Figure 4.16), explain the specific positions of the mitochondrion and chloroplast lineages on this tree==
This tree indicates that the sequences of rRNA and other genes in mitochondria are most closely related to those of proteobacteria, while the sequences of chloroplast genes are most closely related to those of cyanobacteria. These gene sequence relationships are what would be predicted from endosymbiont theory, which posits that both mitochondria and chloroplasts originated as engulfed prokaryotic cells
The first major split in the history of life occurred when ___ diverged from other organisms
- bacteria
]]Horizontal Gene Transfer:]]
The transfer of genes from one genome to another genome through mechanisms such as transposable elements, plasmid exchange, viral activist, and perhaps fusions of different organisms
A tangled web of life - Horizontal gene transfer may have been so common in the early history of life that the base of a “tree of life” might be more accurately portrayed as a tangled web
]]Carotenoids:]]
Colored molecules that have diverse functions in many organisms, such as photosynthesis in plants and light detection in animals
]]Acyrthosiphon pisum (Pea Aphid):]]
A small plant-dwelling insect whose genome includes a full set of genes for the enzymes needed to make carotenoids (animals generally cannot synthesize carotenoids from scratch)
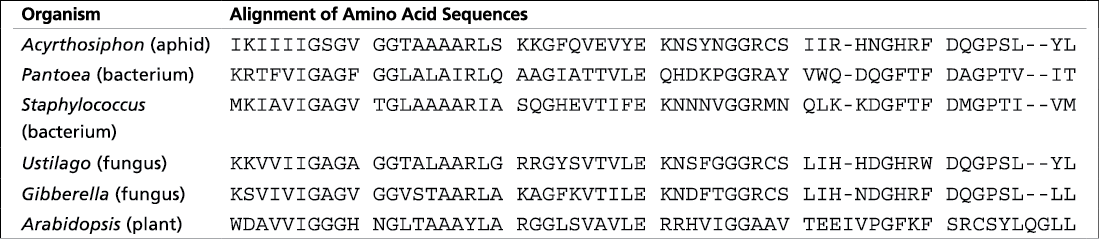
Hypothesize how aphids got the genes to synthesize carotenoids
Since animals lack the genes, it likely wasn’t inherited from a single-celled common ancestor shared with microorganisms and plants. It was likely acquired by horizontal gene transfer from distantly related organisms
The sequences below show the first 60 amino acids of one polypeptide of the carotenoid-biosynthesis enzymes in the plant Arabidopsis thaliana (bottom) and the corresponding amino acids in five nonplant species, using the one-letter abbreviations for the amino acids. A hyphen (-) indicates a gap inserted in a sequence to optimize its alignment with the corresponding sequence in Arabidopsis
==Why is the kingdom Monera no longer considered a valid taxon?==
The kingdom Monera included bacteria and archaea, but we now know that these organisms are in separate domains. Kingdoms are subsets of domains, so a single kingdom (like Monera) that includes taxa from different domains is not valid
==Explain why phylogenies based on different genes can yield different branching patterns for the tree of all life==
Because of horizontal gene transfer, some genes in eukaryotes are more closely related to bacteria, while others are more closely related to archaea; thus, depending on which genes are used, phylogenetic trees constructed from DNA data can yield conflicting results
==Draw the three possible dichotomously branching trees showing evolutionary relationships for the domains Bacteria, Archaea, and Eukarya. Two of these trees have been supported by genetic data. Is it likely that the third tree might also receive such support?==
The fossil record indicates that prokaryotes originated long before eukaryotes. This suggests that the third tree, in which the eukaryotic lineage diverged first, is not accurate and hence is not likely to receive support from genetic data 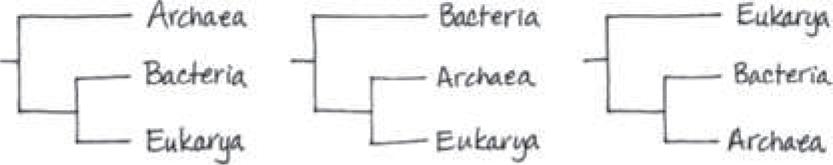
==Why was the five-kingdom system abandoned for a three-domain system?==
Genetic data indicated that many prokaryotes differed as much from each other as they did from eukaryotes. This indicated that organisms should be grouped into three “super-kingdoms,” or domains (Archaea, Bacteria, Eukarya). These data also indicated that the previous kingdom Monera (which had contained all the prokaryotes) did not make biological sense and should be abandoned. Later genetic and morphological data also indicated that the former kingdom Protista (which had primarily contained single-celled organisms) should be abandoned because some protists are more closely related to plants, fungi, or animals than they are to other protists