lecture 9: DNA topoisomerase 1
As DNA unwinds, it acts like a rope. when the strands are pulled apart, twisting tension is added and one twist is added per twist unwound. eventually, the twist can’t be unwound until the tension is removed. This is best characterised in small DNA. One topoisomerase enzyme cuts 1 strand and passes the other strand through the break and joins the break together. this removes one twist and so allows the full DNA to be unwound. Topoisomerase 2 cuts both strands of the double helix, and passes another double strand through this and release this. This means that supercoiling is removed 2 twists at a time. Topoisomerases change the shape dynamics of DNA. supercoiling exists to pack all of the DNA into individual cells. These enzymes fine tune the steady state structure of DNA to allow protein interactions with the DNA and to prevent excessive supercoiling. These enzymes are also medically important as they are target of important drugs such as anti-cancer drugs and antibiotics. cleavage of phosphodiester bonds in all cases are accomplished by nucleophilic attack from specific tyrosine resides within each topoisomerase. Tyrosine has a hydroxyl group on an aromatic ring. The hydroxyl can ionise and form a covalent link to DNA. the pKa of tyrosine in the local environment of these enzymes allows ionisation
In an agarose gel, DNA further down the gel is highly supercoiled and the DNA at the top is relaxed. If the DNA is treated with a topoisomerase, gradual levels of relaxation can be seen going up the gel. These enzymes achieve this thrugh making covalent linkages that are broken. In cells therefore, you can have different packaged forms such as knots. Topoisomerases are also capable of solving these.
In DNA cleavage, you can have attack of the tyrosine on the topoisomerase onto the phosphate bond between bases. This can attach to the 5’ end or to the3’ end and this determines what type the topoisomerase is. There can be the Type 1 enzymes that break one strand or the type 2 that break 2
the type 1 topoisomerases are large but single polypeptides. If it is a type 1a, there is a 5’ linkage formed and type 1b form 3’ linkage. Viruses can also have topoisomerase type enzymes.
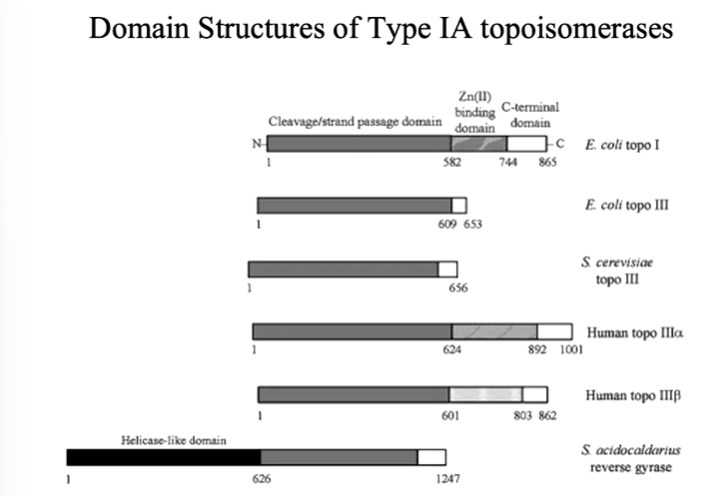
The C terminus usually has the active site tyrosine. Type 1a enzymes are in general monomeric and cleavage of DNA stand is accompanied by covalent attachment of one DNA ends through a 5’ phosphodiester bond to an active site tyrosine. They require metal ion for relaxation activity - usually Mg2+. Plasmids with negative but not positive supercoils are substrates for type 1a topoisomerases and relaxation of negative supercoils does not go to completion. Relaxation of the DNA change the linking number by steps of 1.
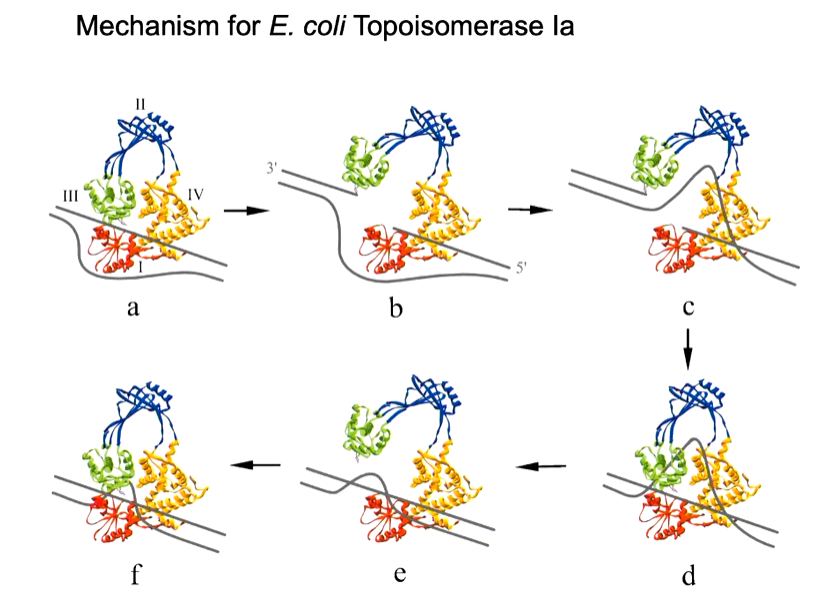
mechanism:
bind a double strand of part of DNA
induce in that strain and unwinding
make a break in one strand using tyrosine residue
unbroken strand passes through gap - conformational change
reformation of the double strand
This is inferred through many structures found that describe this. Human type 1b enzyme bound to DNA was the first structure in the presence of DNA. The human protein was expressed and purified as an N terminal truncated but active form was missing - the first 174 amino acids. Only the density for residues after residue 215 is visible for entire N terminal domain view was missing. The DNA protein interaction extended over 14 bps with the majority of points of contacts 5 bps upstream to the cleavage site. The protein was inactivated by mutation of the active site tyrosine to phenylalanine. This means that DNA can bind but there is no cleavage of the phosphodiester bond.
Type 2 topoisomerases produce double strand breaks in part of a DNA strand and pass another region through that break. This process is ATP dependant. IN the case of bacterial enzymes, there is a protomer that is split into 2. These enzymes are large.
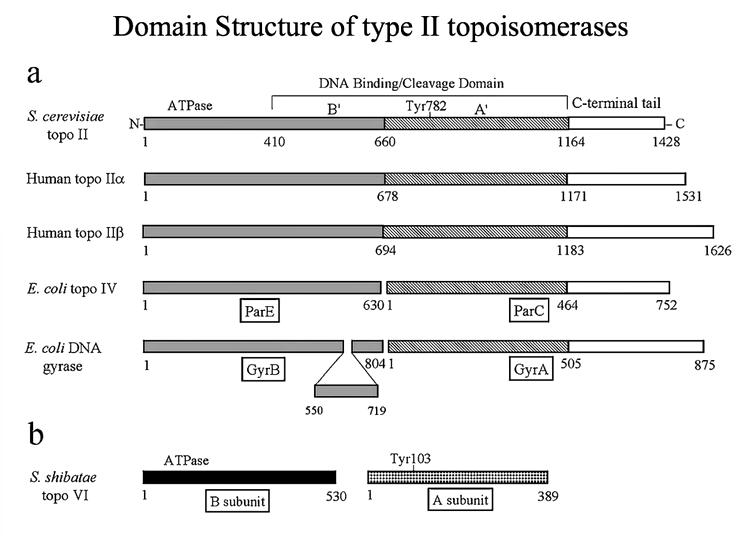
The dimeric enzymes bind duplex DNA and cleave the opposing strands with a 4 bp stagger - not to be confused with restriction enzymes that cuts both strands but there is no defined recognition sequence here. Cleavage of the DNA is via a tyrosine residue to the 5’ of the DNA that makes a phosphotyrosine bond. Conformational changes pull the two cleaved ends of the DNA apart to create an opening for strad passage. The opening is referred to as the gated or G segment DNA whereas the strand that moves through the gate is referred to as the transported or T segment. These reactions are ATP (Mg2+) dependent although one cycle of relaxation can occur in the presence of a non hydrolysable form of ATP for DNA gyrase.
The linking number changes in steps of 2. Bacterial topoisomerases (gyrases) have ATPases and DNA binding/cleavage domains in separate proteins and are A2B2. Eukaryotic topos are single chain but dimeric. Structural information from several topo II strains have revealed that the active site tyrosine lies in a HTH motif.

One of the first structures of Topo II of a part of DNA gyrase - N terminal fragment of E coli GyrB. The N terminal section comes into a major region with the ATP binding site or non hydrolysable form). This is a dimer and the N terminus of one protomer reaches over and forms part of the ATP binding site of the other protomer. The ATP binding site is made up of both these sites. There is a big hole in this structure where the DNA goes in. The active site tyrosine is also in this region between two sides in the dimer structure. There is an interface that forms a cavity where DNA binds. A composite structure of topo II formed from the amino terminal ATP binding domain of E Coli topo II and the carboxyl terminal fragment from yeast topo II show an N gate and a C gate:
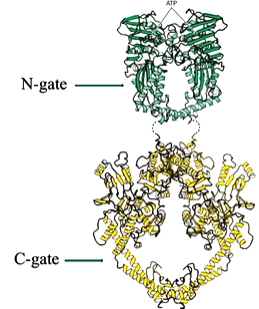
This structure led to the two gate model for Topo Ii mechanism
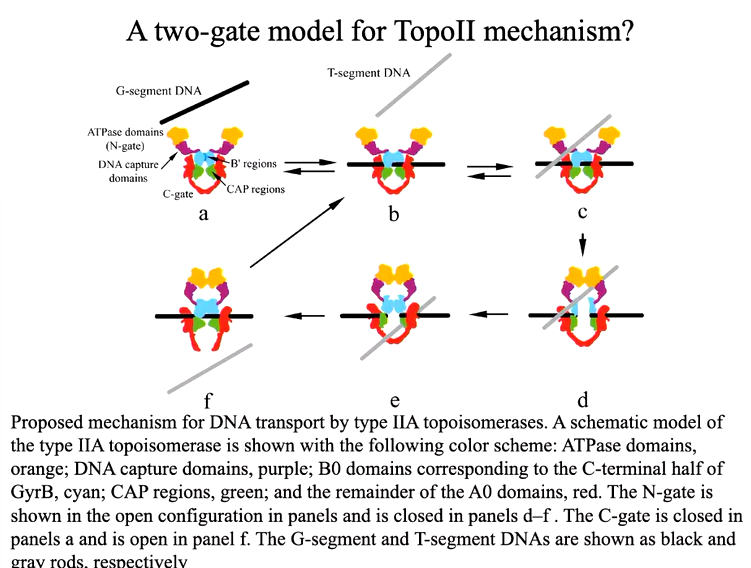
Whilst parts of either eukaryotic topo IIs and bacterial gyrases have been crystalised and structured determined, we do not have a clear idea of how the domains fit together to produce the structure function relationship that is present for biological activity. One of the central mysteries around Topo Ii structure and function concerns the mechanisms through which nucleotide binding, hydrolysis and release and transduced unto gate opening and closure.