Transcription in Eukaryotes (Lectures 9-12)
RNA Polymerases
Prokaryotes: There is one RNA polymerase
Eukaryotes: 3 distinct RNA polymerases with specialized binding affinity for different classes of genes
Pol 1
A distinct RNA polymerase in eukaryotes with a binding affinity for major ribosomal RNAs (rRNA: 5.8S, 18S, 28S)
Makes large rRNA precursor in nucleolus
Pol II
A distinct RNA polymerase in eukaryotes with a binding affinity for genes encoding proteins
Makes heterogeneous nuclear RNA (hnRNA) and small nuclear RNA
Pol III
A distinct RNA polymerase in eukaryotes with a binding affinity for genes encoding 5S rRNA, tRNAs (for translation), small nuclear RNAs for RNA splicing
Cis-acting elements
Regulatory DNA sequences on themselves, regulation by the Sam molecule that is being regulated
Trans-acting elements
Regulation is by a different molecule that is being regulated
enhancers: activators bind and its affects transcription, often far way, in another gene
Alpha-amanitin
A toxic cyclic peptide found in mushrooms
An inhibitor for Pol II and III so will no longer create essential RNAs, showing why certain mushrooms are toxic and deadly
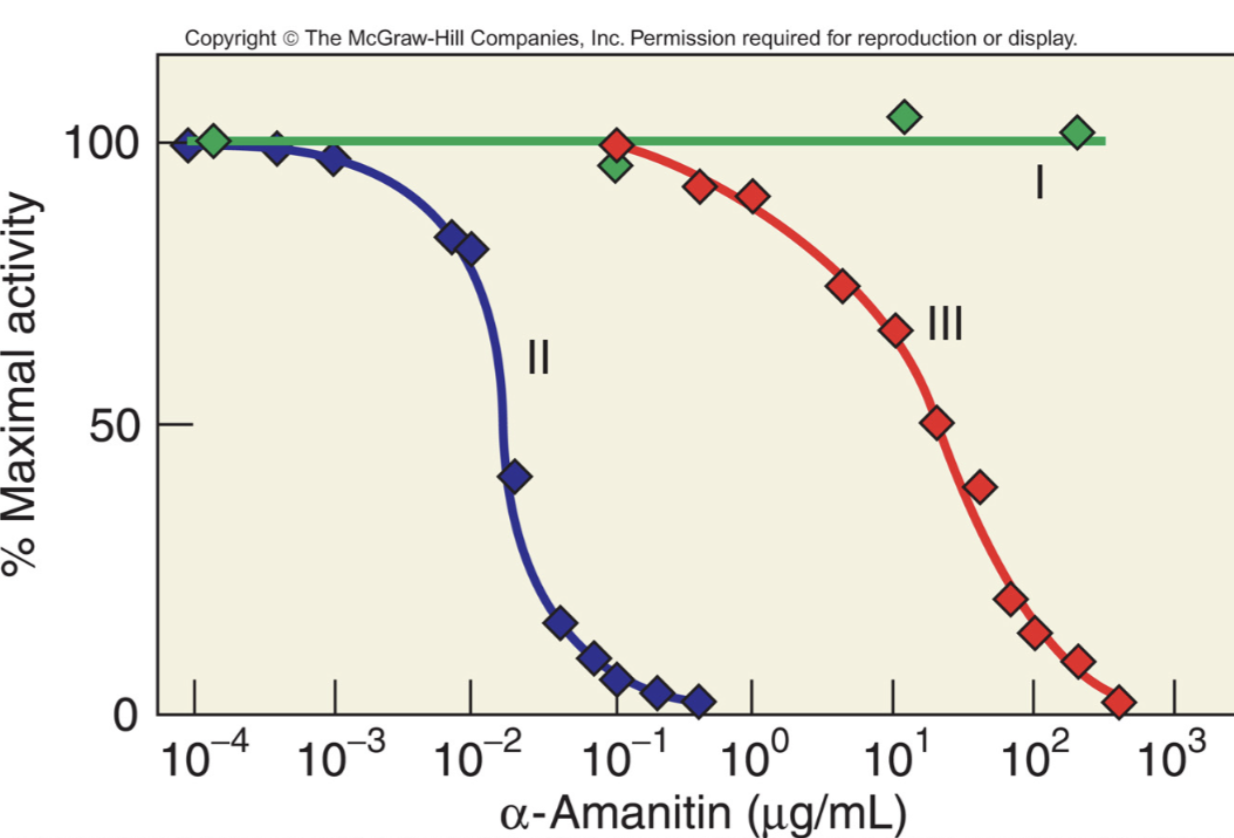
Image shows that even with very small amounts, activity for Pol II and III is greatly limited
Pol II Structure
12 subunits and has been sequences in yeast
3 subunits resemble the core subunits of bacterial RNA polymerases in both structure and function
5 are found in all three nuclear RNA polymerases, 2 are not required for activit and 2 fall into none of these categories
How were the RNA Polymerase subunits tagged?
A epitope tag is genetically added to one subunit of the yeast RNA polymerase while all other subunits are normal, forming an active polymerase
An antiepitope antibody is used against the tag, which immunoprecipitates the whole RNA polymerase
Gel electrophoresis separates the denatured subunits
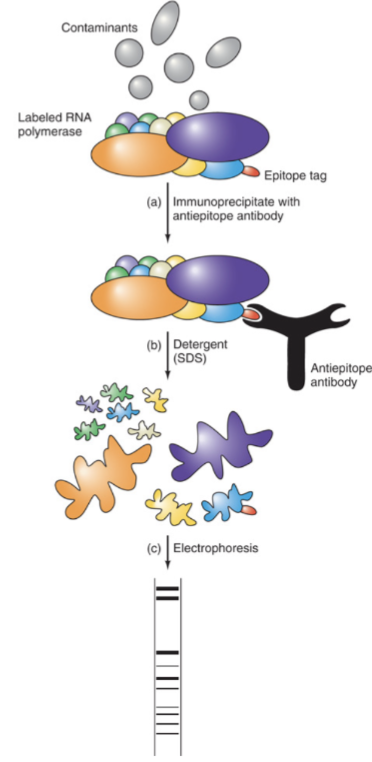
What are the different roles of Rpb1 subunit?
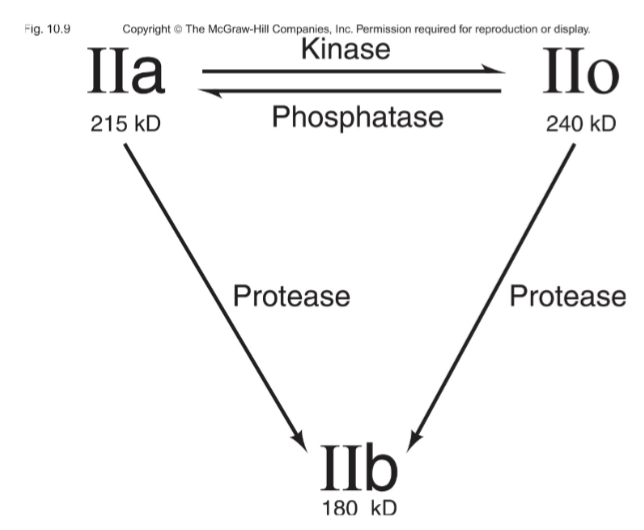
Gene that encodes that largest subunit of Pol II
Subunit IIa is the primary product in yeast but can be converted to:
IIb by the removal of CTD (7-peptide repeat)
IIo through phosphorylating 2 serine in the repeated heptad of the CTD
IIa binds to the promoter
IIo is involved in transcription elongation
Enhancers/Silencers
Distal elements, far from start, ± 50 kb from start
Position and orientation independent DNA elements that stimulate or depress
Often tissue specific; rely on tissue specific DNA-binding proteins for their activities
Some DNA elements can act either as a enhancer or silencer depending on what it is bound to
Promoter
Proximal element, upstream of start, -200 to -40 bp
Helps attract general transcription start site and direction of transcription
Core promoter
Very close to the starting point, minimal region required fro accurate initiation of transcription
Attracts general transcription factors and RNA polymerase II at a basal level and sets the transcription start site and direction of transcription
Modular and can contain almost any combo of the following:
TATA box
TFIIB recognition element (BRE)
Initiator (Inr)
Downstream promoter element (DPE)
Downstream core element (DCE)
Motif ten element (MTE)
TATA-less promoters tend to have DPEs
Highly specialized genes tend to have TATA boxes
Class II promoters
Recognized by RNA polymerase II
Core promoter
Proximal promoter
Enhancer
Act through proteins that are bound to them; called activators
Stimulate transcription by interacting with other proteins called general transcription factors at the promoter that promote the formation of a pre initiation complex
Frequently found upstream of the promoter (not always though)
Silencers
DNA elements that can inhibit (at a distance) transcription
They work by causing the chromatin to coil up into a condensed, inaccessible and inactive form preventing the transcription of neighboring genes
Transcription in Eukaryotes
RNA polymerases are incapable of binding themselves to their promoters so they rely on proteins called transcription actors to guide them
general transcription factors and gene-specific transcription factors (activators
General transcription factors
combine with RNA polymerase to form a pre-initiation complex that is able to initiate transcription when nucleotides are available
tight binding involves formation of an open promoter complex with DNA at the transcription start site that has melted
DABpolFEH (Class II Pre-initiation) Complex
RNA polymerase II
General transcription factors: TFIIA, TFIIB, TFIID, TFIIF, TFIIE, TFIIH
TF and polymerase bind in a specific order
Order of TF binding in DABpolFEH
TFIID with TFIIA binds to the TATA box forming the DA complex
TFIIB binds next, generating the DAB complex
TFIIF helps RNA polymerase bind to a region -34 to +17 bp, forming the DABpolF complex
The TFIIE and TFIIH bind to form the complete pre-initiation complex DABPolFEH
In vitro- participation of TFIIA seems to be optional
Structure and Function of TFIID
TATA-box binding proteins (TBP)
Highly evolutionarily conserved
Binds to the minor groove of the TATA box
Saddle-shaped TBP lines up with DNA, underside of the saddle forces open the minor groove
TATA box is bent into 80° curve
TBP-associated factors (TAFs) specific for class II
What side does TBP bind to TATA box
Inosine (I) and adenine look alike in the the minor grove, but different in major
Thymidine and cytidine look identical from minor groove
Use of TBP
Universal transcription factor required by Class I, II, III genes
TBP mutant cells do not transcribe these genes
Require transcription of at least some genes of Archaea, single celled organisms lacking nuclei
TAF1, TAF2
TBP-Associated Factors
Help the TFIID bind to the initator of the DPE of promoters
Enable TBP to bind to TATA-less promoters that contain elements such as GC box
Enzymatic activities of TAF1
Histone acetyletransferase (HAT)
Protein kinase
Structure and Function of TFIIB
Binds to TBP at the TATA box via C-terminal domain and polymerase II via its N-terminal domain
Important role in establishing transcription start site
TFIIH
Last TF to going pre-initiation complex
Structure
4 subunits: protein kinase
5 subunits: Core with 2 DNA helicase
Roles
Phosphorylates CTD of RNA polymerase II
Unwinds DNA at the transcription start site to create transcription bubble
Expansion releases the stalled RNAPII and allows it to clear the promoter
How is IIO (phosphorylated form of polymerase enzyme) created?
Pre-initiation complex forms with hypophosphorylated form of RNA polymerase II (IIA)
TFIIH phosphorylates serine 5 in CTD (carboxyl-terminus domain (Tyr-Ser-Pro-Thr-Ser-Pro-Ser)) of the largest subunit creating the phosphorylated form
Essential for initiation of transcription
How does serine phosphorylation change when there is a shift from initiation to elongation?
transcription: TFIIH phosphorylates serine 5
elongation: CTDK-1 phosphorylates serine 2
Activators
Gene-specific transcription factors that bind to enhancers to provide an extra needed boost to transcription
Also recruit RNA polymerase to promoters
Stimulate binding of general transcription factor and RNA polymerase to a promoter
TF Protein structure
DNA binding domain
Activation/repression domain
Often dimerization domain
Sometimes binding sites for hormones that regulate the TF
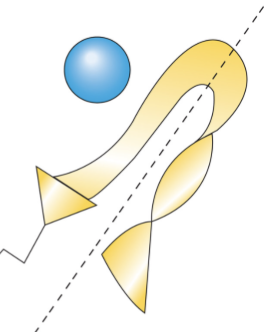
Zinc Fingers
Type of TF
An antiparallel beta-sheet and an alpha-helix
Two cysteines in the beta sheet and two histidines in the alpha-helix coordinate the zinc ion in the middle
Zinc does not bind to DNA
Gal4 DNA binding
yeast activator
Has a zinc finger in the first 60 aa to coordinate DNA binding
Contains a DNA-binding motif with six cysteines that complex two zinc ions
each monomer contains dimerization (two molecules combine to form a larger molecule called a dimer) domain
bZIP (leucine zipper) transcription factor and bHLH
One part of the domain contains a region that mediates sequence specific DNA binding properties and the leucine zipper that is required to hold together (dimerize) two DNA binding regions.
Binding sites in major grove of DNA
Must have another domain to bind RNA polymerase
Leucine on hydrophobic faces of helices interact to enable dimerization
Hypotheses for RNA polymerase recruitment to promoter
General TF causes a stepwise build-up of pre-initiation complex
General TF and other proteins are already bound to polymerase in a complex called RNA polymerase holoenzyme
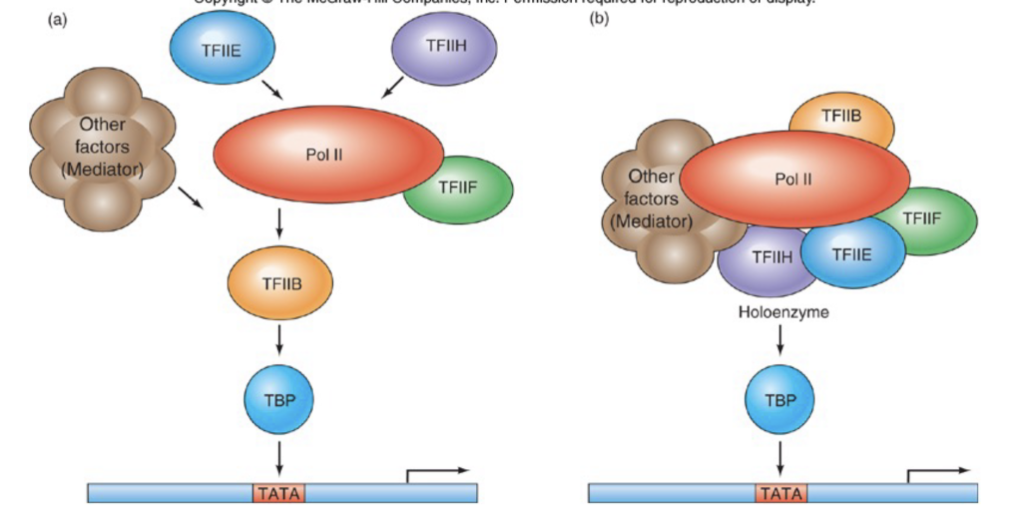
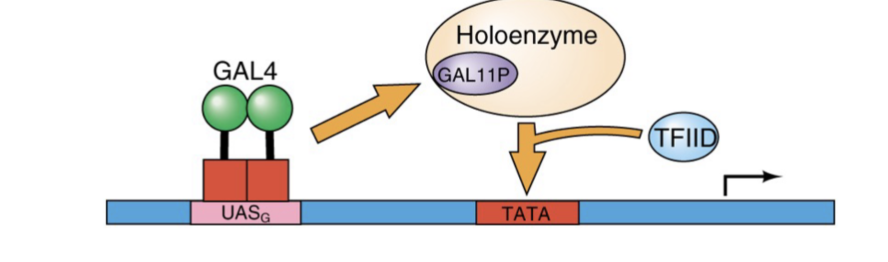
Hypothesis #2 model using GAL11P-containing holoenzyme
Dimerization domain of GAL4 binds to GALL11P in holoenzyme
After dimerization, holoenzyme and TFIID binding to the promoter
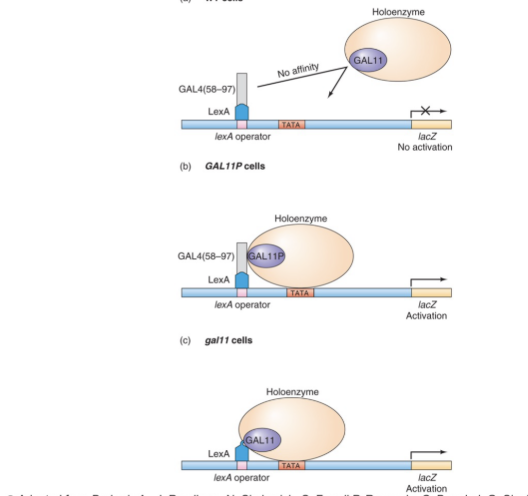
this would not work with GALL11 as it does not bind to GAL4 like GAL11P does
instead LexA DNA binding to GAL11 mimics transcriptional activation activity of GAL11P
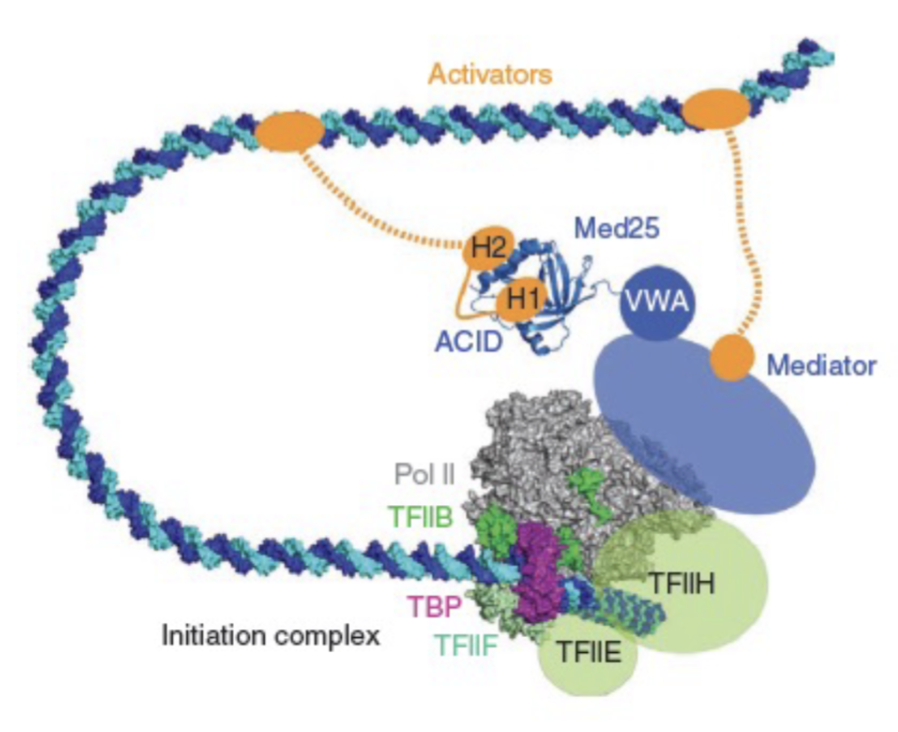
Whole process of gene activation by RNA Polymerase II
RNA Polymerase II is recruited to form the pre-initation complex
Thousands of base pairs upstream, an activator bound to an enhancer loops to activate the RNA polymerase
CTD is phosphorylating, initiating transcription
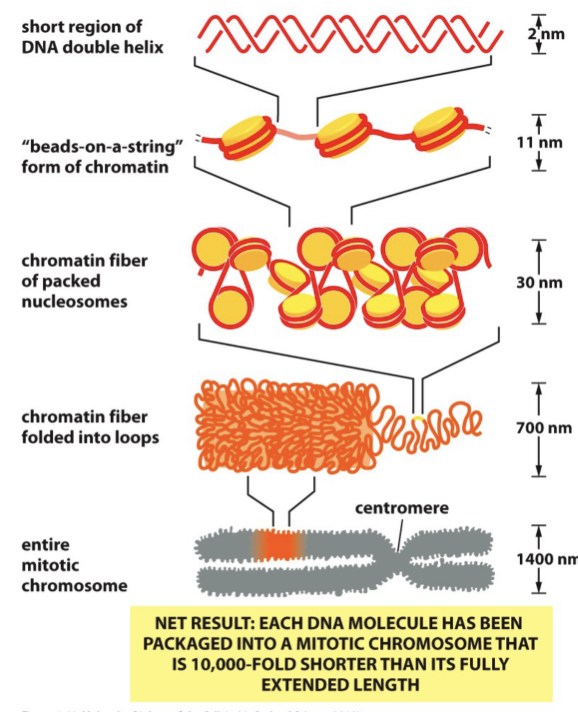
Chromatin Structure
Composed of DNA and proteins, histones
Roughly equal masses of DNA and histones
1 histone octamer (8 histone proteins (2 of H2A, H2B, H3, H4into a disk)) per 200 bp of DNA
Beads on a string form a chromatin
Chromatin Packing
Many levels of chromatin packing postulated to give rise to the highly conensed mitotic chromosome
Core histone structure
Histones 2A and 2B form a dimer, H3 and 4 form a dimer
Bending of DNA in a nucleosome (the histone and DNA)
DNA makes 1.7 tight turns around the histone
Minor groove is compressed on the inside of the turn
142 hydrogen bonds are formed between DNA and the histone core in each nucleosome
Role of histone H1
Binds to each nucleosome, contacting both DNA and protein, changing the path of the DNA as it exits from the nucleosome to help compact nucleosomal DNA
Histone impact on transcription
Modifications to histones can impact transcription by making DNA more or less accessible to proteins
DNA can be too tightly wrapped for proteins to access
Nucleosome sliding
Allows proteins to find targets faster
Catalyzed by ATP-dependent chromatin remodeling complexes (ISW1 or SWI)
Uses energy from ATP hydrolysis to push on DNA and loosen attachment to nucleosome core
read-writer complex can spread chromatin change along a chromosome
Histone methylation
Covalent modification of core histone tails
Adds 1,2, or 3 methyl groups to nucleosomal histones to either activate or repress transcription
Typically modifies Lysine
Histone phosphorylation
Covalent modification of core histone tails
Adds a phosphate group
Histone acetylation
Covalent modification of core histone tails
Acetyl groups are added to lysine residues within histone tails, a key epigenetic modification that loosens chromatin structure and promotes gene expression
Histone Code
Hypothesis proposing that specific patterns of chemical modifications (post-translational modifications or PTMs) on histone proteins
Histone writers
Proteins that add histone markers
Histone acetlytransferase
Histone erasers
Proteins that remove histone marks
Histone deacetylase
Histone reader proteins
Recognizes Histone modification marks
Binds when a certain mark is present
Can be involved in chromatin remodeling
Nucleosome displacement/relocation
Histone replacements
Model for histone code at human IFN-B promoter
IFN-β is an inflammatory cytokine
Activators recruit GCN5 (histone acetyltransferase) to acetylate H4K8 and H3K9
Activators recruit a kinase to phosphorylate H3S10
H4K8 attracts SWI/SNF (nucleosome slider) to remodel nucleosome
Now allows the binding of TFIID which is now attracted by the TATA box but also H3K9ac and H3K14