Molecular Genetics EXAM 3 (copy)
Recombination CH13
Introduction
Site specific recombination involves specific DNA sequences
Somatic Recombination- recombination that occurs in nongerm cells (not during meiosis); most commonly used to refer to recombination in the immune system
recombination systems have been adapted for experimental use
Homologous Recombination Occurs Between Synapsed Chromosomes in Meiosis
Chromosomes must synapse (pair) in order for chiasmata to form where crossing over occurs
the stages of meiosis can be correlated with the molecular events at the DNA level
sister chromatid- each of two identical copies of a replicated chromosome; this term is used as long as the two copies remain linked at the centromere
sister chromatids separate during anaphase in mitosis or anaphase II in meiosis
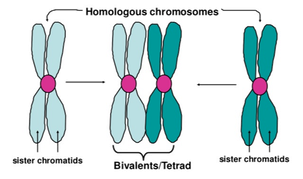
bivalent- the structure containing all four chromatids (two representing each homolog) at the start of meiosis
a sister chromatid from mom and sister chromatid from dad makes a bivalent
synaptonemal complex- the morphological structure of synapsed chromosomes
joint molecule- a pair of DNA duplexes are connected together through a reciprocal exchange of genetic material
Double Strand Breaks Initiate Recombination
The double strand break repair (DSBR) model of recombination is initiated by making a double strand break in one (recipient) DNA duplex and is relevant for meiotic and mitotic homologous recombination
in 5’ end resection, exonuclease action generates 3’-single-stranded ends that invade the other (donor) duplex
when a singe strand from one duplex displaces its counterpart in the other duplex (single-strand invasion), it creates a branched structure called a D-loop
strand exchange generates a stretch of heteroduplex DNA consisting of one strand from each parent
New DNA synthesis replaces the material that has been degraded
branch migration- the ability of DNA strand partially paired with its complement in a duplex to extend its pairing by displacing the resident strand with which it is homologous
Capture of the second double-strand break end by annealing generates a recombinant joint molecule in which the two DNA duplexes are connected by heteroduplex DNA and two Holliday junctions.
The joint molecule is resolved into two separate duplex molecules by nicking two of the connecting strands.
Whether recombinants are formed depends on if the strands involved in the original exchange or the other pair of strands is nicked during resolution
Gene Conversion Accounts for Interallelic Recombination
Heteroduplex DNA that is created by recombination can have mismatched sequences where the recombining alleles are not identical
Repair systems may remove mismatches by changing one of the strands so its sequence is complementary to the other
Mismatch (gap) repair of heteroduplex DNA involves a process that recognizes the incorrect base pairing and facilitates the correction of these mismatches through various repair mechanisms, ultimately leading to gene conversion and increased genetic diversity.
Break-Induced Replication Can Repair Double-Strand Breaks
Break-induced replication (BIR) is initiated by a one-ended double-strand break
BIR at repeated sequences can result in translocations
Recombining Meiotic Chromosomes Are Connected by
the Synaptonemal Complex
During the early part of meiosis, homologous chromosomes are paired in the synaptonemal complex
the mass of chromatin of each homolog is separated from the other by a proteinaceous complex
axial element- a proteinaceous structure around which the chromosomes condense at the start of synapsis
lateral element- a structure in the synaptonemal complex that forms when a pair of sister chromatids condenses on to an axial element
central element- a structure that lies in the middle of the synatonemal complex, along which the lateral elements of homologous chromosomes align
it is formed from zip proteins
recombination nodules (nodes)- dense objects present on the synaptonemal complex; they may represent protein complexes involved in crossing over
Specialized Recombination Involves Specific Sites
specialized recombination involves reaction between specific sites that are not necessarily homologous
recombinase- enzyme that catalyzes site-specific recombination
phage lambda integrates into the bacterial chromosome by recombination btwn the attP site on the phage and the attB site on the E. coli chromosome
core sequence- the segment of DNA that is common to the attachment sites on both the phage lambda and bacterial genomes
it is the location of the recombination event that allows phage lambda to integrate
The phage is excised from the chromosome by recombination between the sites at the end of the linear prophage.
Phage lambda int encodes an integrase that catalyzes the integration reaction.
Integrases are related to topoisomerases, and the recombination rxn resembles topoisomerase action except that nicked strands from different duplexes are sealed together
Recombination Pathways Adapted for Experimental Systems
homologous recombination allows for targeted transformation
The Cre/lox and Flp/FRT systems allow for targeted recombination and gene knockout construction.
Linkage Mapping CH12
Linkage and Crossing over
crossing over may produce recombinant genotypes
crossing over may alter the linkage of genes
bivalent chromosomes consist of two homologous chromosomes with a pair of sister chromatids each
genetic recombination by crossing over can produce new combinations of alleles on chromosomes
the process of this leading to a new combination of alleles
the cells that contain the new allelic combinations are called nonparental or recombinant cells
the cells that contain the original combination of alleles are called parental
Bateson and Punnett discovered two traits that did not assort independently
demonstrated that not all traits assort independently
Morgan- the linkage of X-linked genes and proposed that crossing over between X chromosomes can occur
A Chi square analysis- distinguish between linkage and independent assortment
Creighton and McClintock- crossing over produced new combinations of alleles and resulted in the exchange of segments between homologous chromosomes
Crossing over occasionally occurs during mitosis
Chromosomes consist of more than one gene, typically in the hundreds to thousands.
The term linkage is use to indicate:
two genes that are located on the same chromosome.
Genes that are close together tend to be transmitted from parent to offspring as a group
Chromosomes are often called linkage groups, since the genes on a chromosome are physically connected to one another.
Genes that are far apart on a chromosome may assort independently due to crossing over.
In humans there are 22 autosomal linkage groups, the X linkage group, and the Y linkage group.
When geneticists follow different traits in a cross they rely upon dihybrid (two-factor) and trihybrid (three- factor) crosses and so on.
The outcome of the cross depends on whether the genes are linked on the same chromosome or not
Genetic Mapping in Plants and Animals
Background
the frequency of recombination between two genes can be correlated with their map distance along a chromosomes.
Alfred Sturtevant used the frequency of crossing over in dihybrid crosses to produce the first genetic map.
Trihybrid Crosses can be used to determine the order of distance between linked genes.
Interference can influence the number of double crosses over that occur in a short region
Genetic Mapping in Haploid Eukaryotes
Background
Ordered tetrad analysis can be used to map the distance between a gene and the centromere.
Unordered tetrad analysis can be used to map genes in dihybrid crosses
Molecular Evolution
orthologous genes (orthologs) – Related genes in different species.
The minimum size of the proteome can be estimated from the number of types of genes.
Only 1% of the human genome consists of exons.
The exons comprise about 5% of each gene, so genes (exons plus introns) comprise about 25% of the genome.
The human genome has about 20,000 genes
There is no clear correlation between genome size and genetic complexity.
C-value – The total amount of DNA in the genome (per haploid set of chromosomes)
C-value paradox – The lack of relationship between the DNA content (C-value) of an organism and its coding potential.
junk DNA– Non-coding regions of DNA that do not encode proteins but may play roles in regulation and genome stability.
There is an increase in the minimum genome size associated with organisms of increasing complexity.
There are wide variations in the genome sizes of organisms within many taxonomic groups
Roughly 60% of human genes are alternatively spliced.
Up to 80% of the alternative splices change protein sequence, so the human proteome has 50,000 to 60,000 members
monocistronic mRNA – mRNA that encodes one polypeptide.
polycistronic mRNA – mRNA that includes coding regions representing more than one gene
Repeated sequences (present in more than one copy) account for more than 50% of the human genome.
The great bulk of repeated sequences consist of copies of nonfunctional transposons.
There are many duplications of large chromosome regions
Orthologous sequences are genes in different species that evolved from a common ancestral gene through speciation, often retaining similar functions across species.
Paralogous sequences, on the other hand, arise from gene duplication events within the same species, leading to genes that may evolve new functions or acquire different expressions.
Not all genes are essential. In yeast and flies, deletions of less than 50% of the genes have detectable effects.
When two or more genes are redundant, a mutation in any one of them might not have detectable effects
In any particular cell, most genes are expressed at a low level.
scarce (complex) mRNA – mRNA that consists of a large number of individual mRNA species, each present in very few copies per cell.
This accounts for most of the sequence complexity in RNA.
Only a small number of genes, whose products are specialized for the cell type, are highly expressed.
abundance – The average number of mRNA molecules per cell.
abundant mRNA – Consists of a small number of individual species, each present in a large number of copies per cell
mRNAs expressed at low levels overlap extensively when different cell types are compared.
housekeeping gene – A gene that is (theoretically) expressed in all cells because it provides basic functions needed for sustenance of all cell types
The abundantly expressed mRNAs are usually specific for the cell type.
luxury gene – A gene encoding a specialized function (usually) synthesized in large amounts in particular cell types.
About 10,000 expressed genes might be common to most cell types of a multicellular eukaryote
The probability of a mutation is influenced by the likelihood that the particular error will occur and the likelihood that it will be repaired.
synonymous mutation – A change in DNA sequence in a coding region that does not alter the amino acid that is encoded.
nonsynonymous mutation – A change in DNA sequence in a coding region that alters the amino acid that is encoded
In small populations, the frequency of a mutation will change randomly and new mutations are likely to be eliminated by chance.
fixation – The process by which a new allele replaces the allele that was previously predominant in a population
The frequency of a neutral mutation largely depends on genetic drift, the strength of which depends on the size of the population.
The frequency of a mutation that affects phenotype will be influenced by negative or positive selection