Lecture #13: RNA Processing and Measurement
How do RNA strands leave the nucleus?
Through nuclear pores
Movement is mediated by specific transport proteins
Transcription and translation in prokaryotes
Transcription and translation are couples and occur simultaneously in cytoplasm
Termination of RNA synthesis occurs at a specific site; the intact RNA is then translated into proteins on ribosomes
Transcription and translation in eukaryotes
Occur in different cellular locations
mRNA must be transported from the nucleus to cytoplasm before translation occurs
Transport to cytoplasm is also regulated
Termination is not well understood, transcript undergroes several modifications before exporting cytoplasm for translation
Major types of RNA processing
Addition of 5’ CAP
Addition of a POLY A tail
RNA splicing
How mRNA is capped
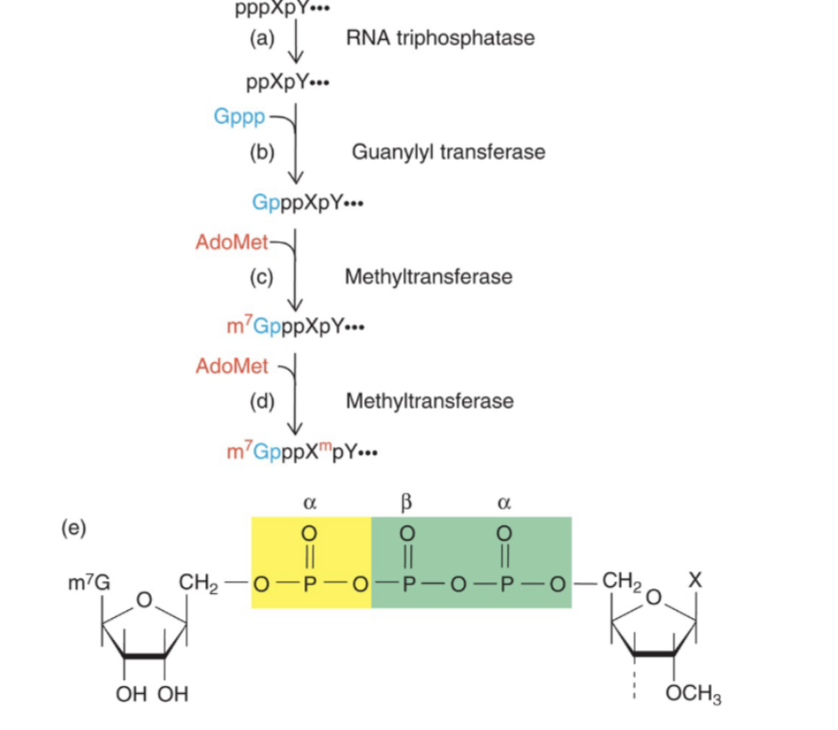
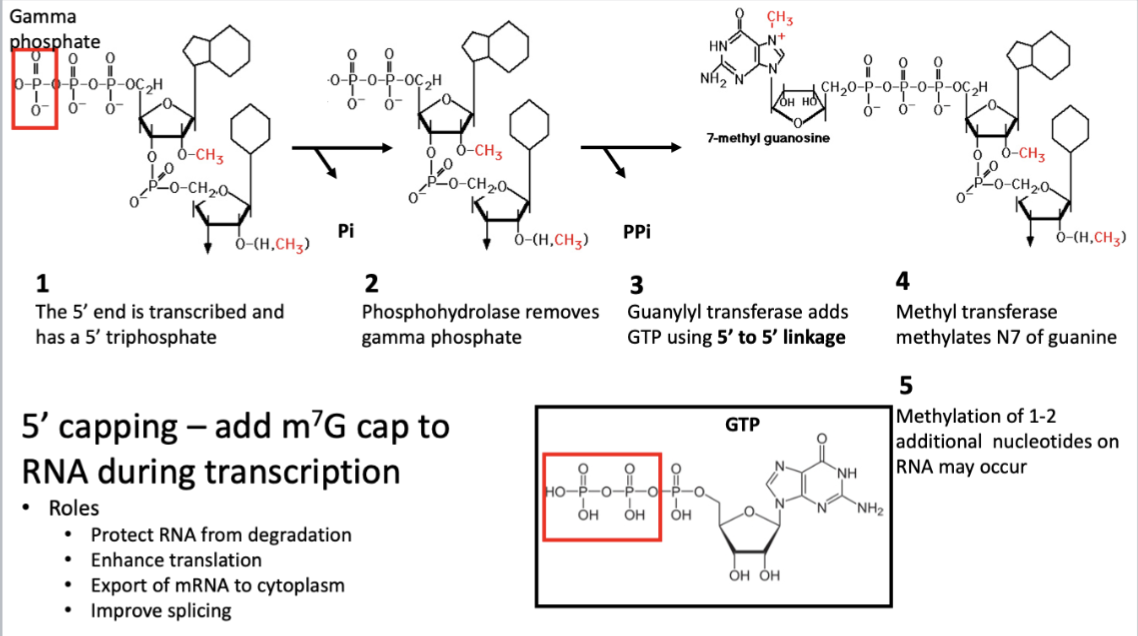
Function of 5’ CAP
enhances transport of mRNA to cytoplasm by transport proteins
enhances translation of mRNA
protects mRNA from degradation
masks 5’ end from exo and endo ribonucleases
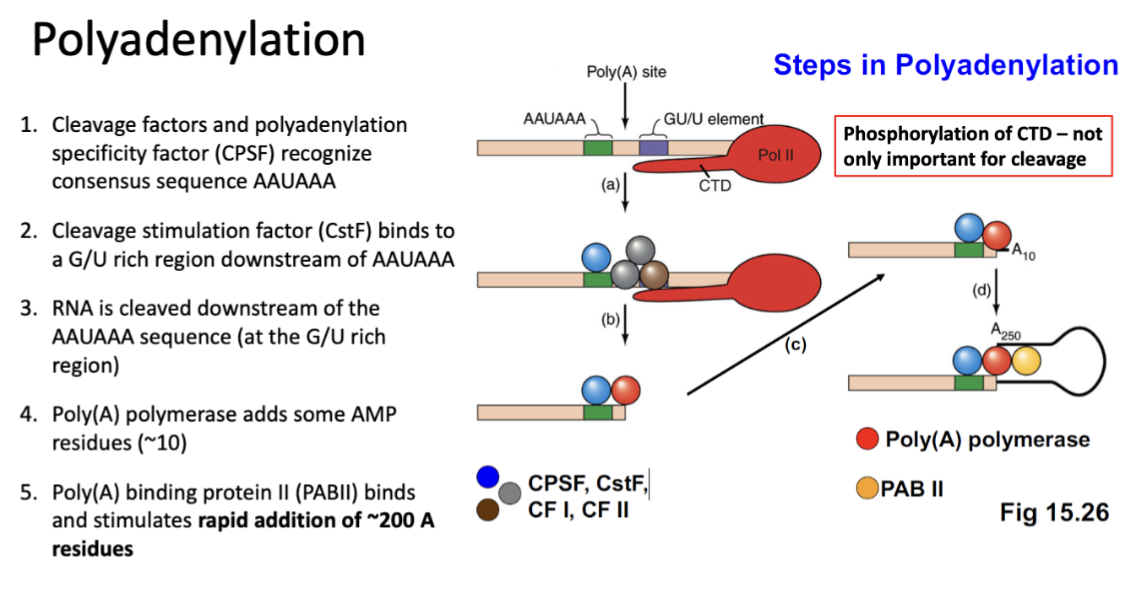
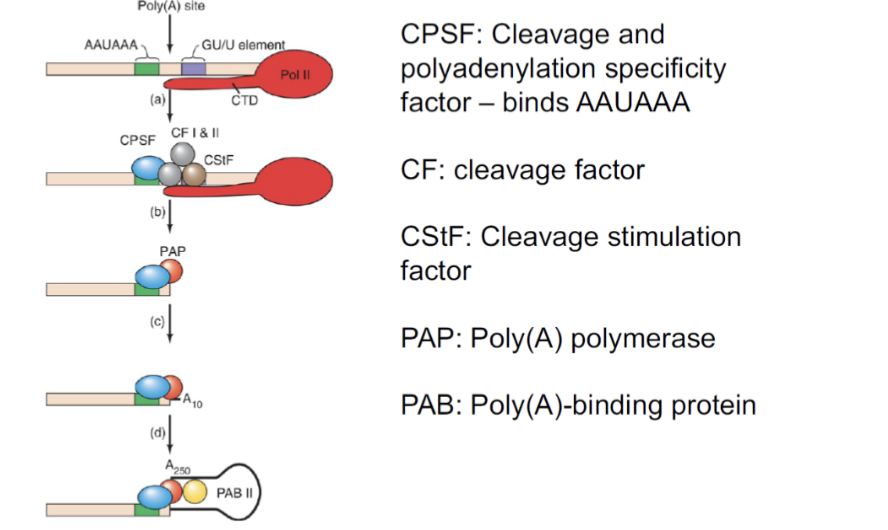
Polyadenylation
Adds 50-250 AMP residues added to 3’ end of transcript
role:
Protect RNA from degradation
Enhances translation of mRNA
Enhances transport of mRNA to cytoplasm by being recognized by specific transport proteins
helps identify RNAs for degradation
RNAs with short polyA tails are more susceptible to RNAses
Steps in polyadenylation
Splicing
Splice introns from primary transcript
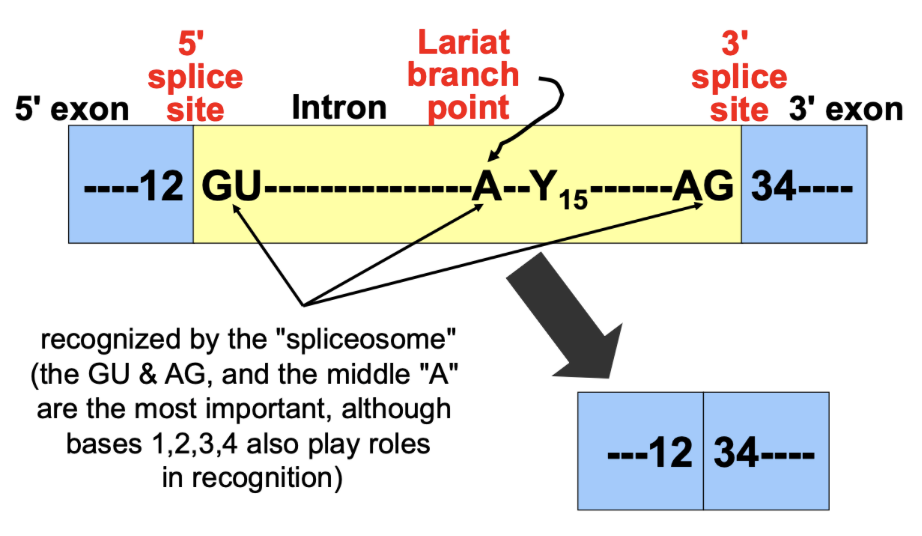
Recognized sequences for splicing
GU and AG are recognized by the spliceosome
A middle “A”: Lariat branch point
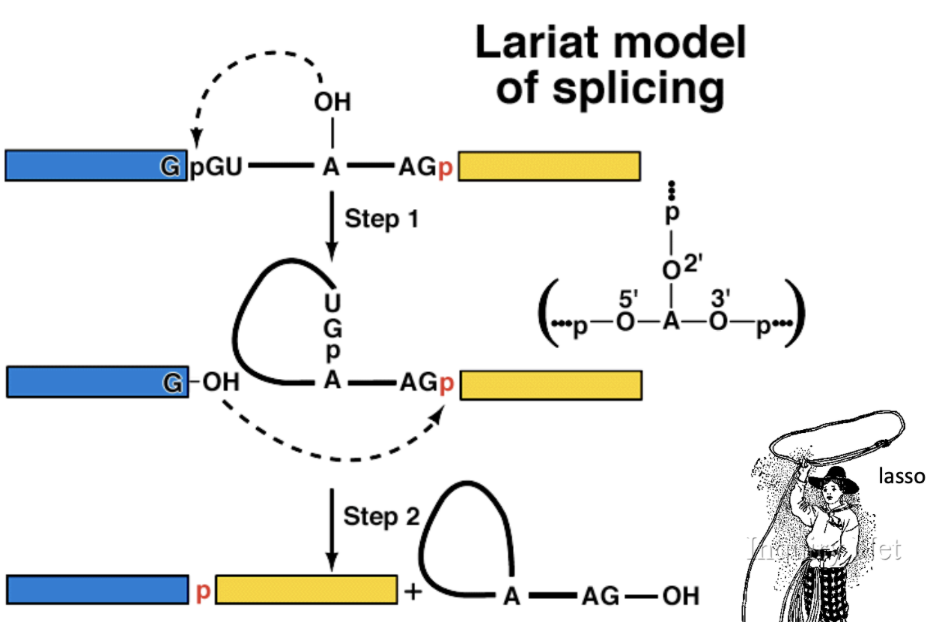
Lariat model of splicing
2’ OH of A (Lariat branch point) attacks phosphodiester bond at 5’ splice site (nucleophilic attack)
Lariat forms by 5’ (g) to 2’ (a)
Free OH on exon 1 attacks phosphodiester bond between intron and exon 2 to cleave 3’ junction
Ligation of exon 1 and exon 2 by formation of a phosphodiester bond→cut is called a lariat which is degraded
Spliceosome is responsible for this process and holds everything together
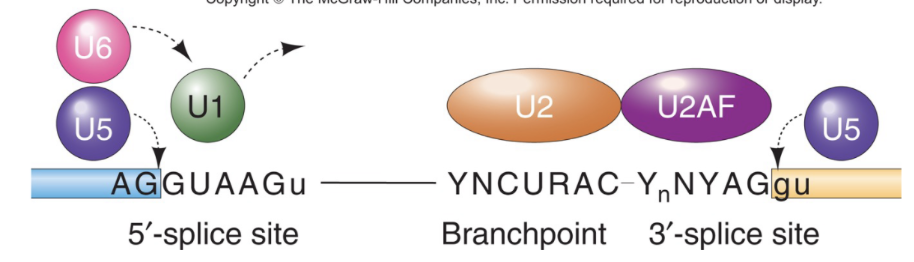
Spliceosome
RNA protein complex with around 70 proteins
snRNPs=Small Nuclear Ribonuclear Proteins
snRNA + Protein =sn RNPs
5 different snurps (U1,2,4,5,6; 3 is used in ribosomal processing)
Steps:
1 recognizes the 5’ splice site first
U1 is replaced by U6
U2 recognizes the branchpoint and protein U2AF (associated factor) recognize the 3’ splice site
A at branch site bulges out and doesnt participate in base pair between snurp and pre-mature RNA
U5 binds to 5’ and 3’ splice sites after initial recognition
Advantages of RNA splicing
Increases # of proteins made form a single gene by alternative splicing (multiple proteins from a single gene)
Regulates gene expression
Introns in DNA bind some transcription factors
What RNAs are effectively transported out of the nucleus?
Ones that are polyadenylated, spliced, and capped
Methods for measuring transcription
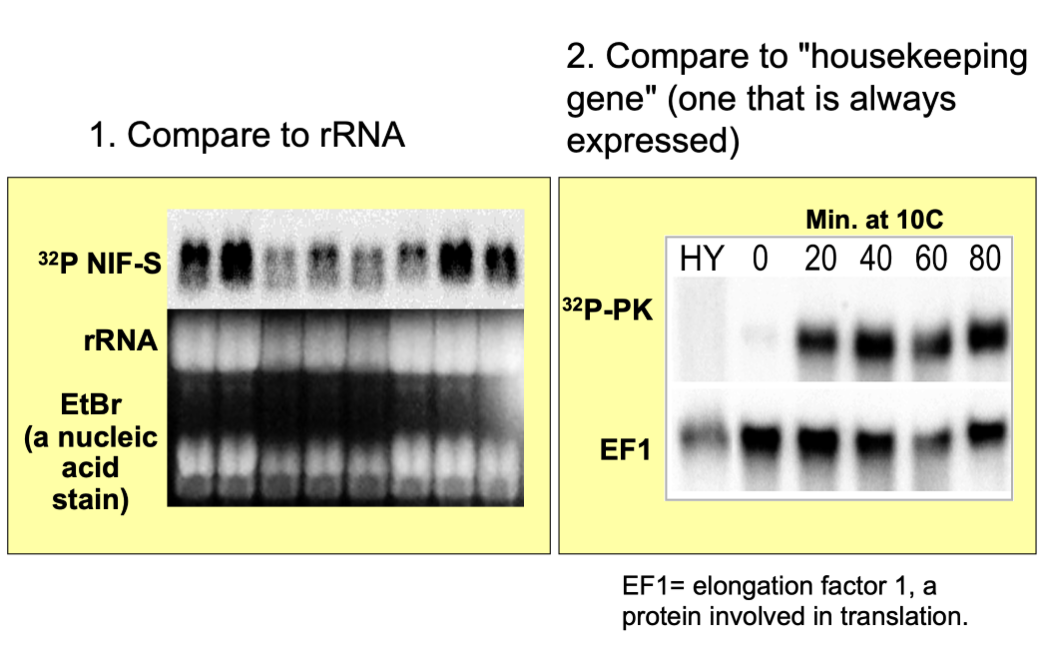
Northern Blots
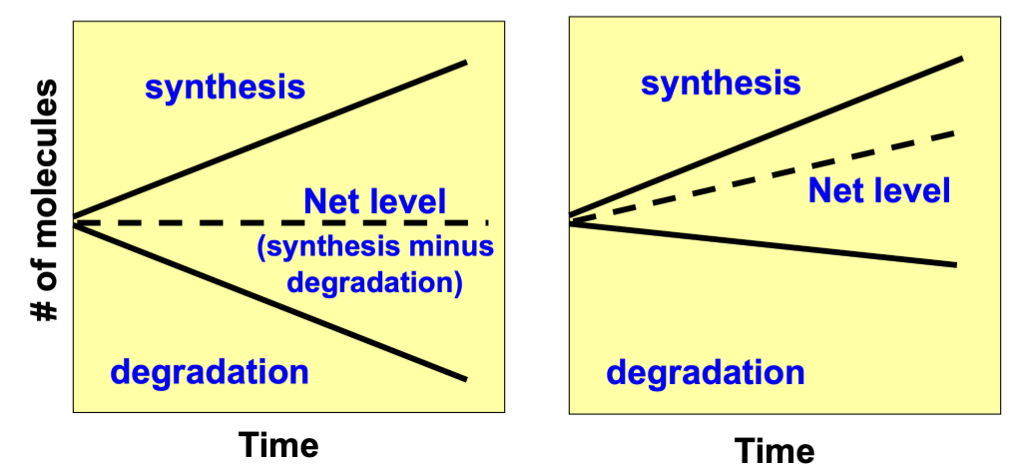
measures RNA abundance
Related to the rate of synthesis versus the rate of degradation
Even if transcription rate is the same, the degradation make be different, leading to differences in Northern blot
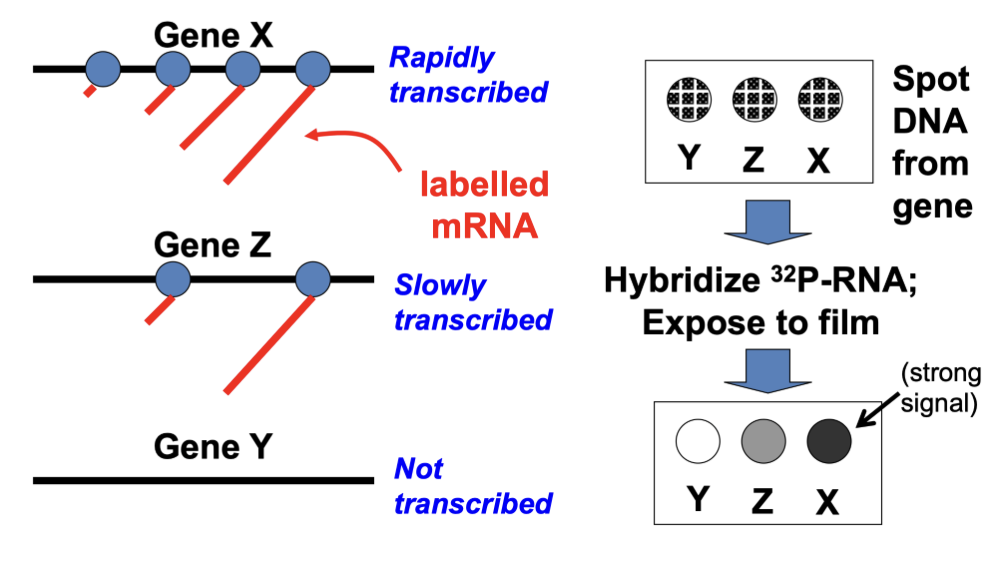
Run-on assays
measures transcription rate (mostly initiation rate)
DNA is isolate
Incubated with a labelled mRNA
Hybridize labelled mRNA is spotted with DNA
The darkness of the color correlated to the transcription rate
each circle is a gene of interest
cDNA
Complementary DNA
Solution to not being able to clone RNA
mRNA can be converted into DNA using reverse transcriptase
RTs: from retrovirus that use RTs to convert their RNA genomes to DNA
Uses of cDNA
express intron-bearing eukaryotic genes in bacteria
compare cDNA and genomic DNA to map introns
measure levels of gene expression
Making cDNA from eukaryotes
Separate mRNA and beads by lowing salt or heat
Convert RNA into DNA using reverse transcriptase and a primer such as oligo-dT
anneal primer
incubate with reverse transcriptase
Reads 3→5 and synthesizes
Add a poly G tail (using terminal transferase)
Remove RNA with RNAse H (specific to DNA/RNA hybrids)
Make dsDNA by adding C primer, then DNA polymerase
Ligate cDNA to linkers containing restriction sites
Digest linkers with restriction enzyme
Ligate to a vector and transform E. coli
Application of cDNA
Make libraries
Compare cDNA versus genomic DNA to define structure of the gene and locations of intron
Produce a protein from an animal gene in bacteria (bacteria cant splice introns