Bio 301 - Ch 9.1 & 10.4 Book Notes
mutations can change genes in many ways
mutations provide an opportunity to trace ancestry and investigate the function of genes through manipulation of the DNA sequence
changes in the DNA sequence of genes by mutation can have a variety of effects on those genes
mutations can change the structure & function of the gene’s products
mutations can also change the regulation of the gene’s expression
gene mutations can have important consequences for the microbe
it may cause additional resistance to antibiotics that normally target the gene’s product
mutation is an important mechanism that can add or subtract genes from genomes
when the codon is being read by the ribosome, the mechanism does not register if bases have been added or removed through mutations
if the number of bases inserted or deleted is not a multiple of three, the reading frame of translation changes
mutations that alter gene sequence fall into several different physical & structural classes
point mutation is a change in a single nucleotide
transition — replacing a purine with a different purine or a pyrimidine with a different pyrimidine
transversion — swapping a purine for a pyrimidine
insertions involve the addition of one or more nucleotides which lengthens the sequence than its original length
serves to alter the reading frame of a DNA sequence
deletions involve the subtraction of one or more nucleotides which shortens the sequence than its original length
serves to alter the reading frame of a DNA sequence
inversion results when a fragment of DNA is flipped in orientation relative to the flanking DNA on either side
duplication produces a second copy of a sequence fragment on the DNA molecule, usually adjacent to the original copy
transposition is the movement of a sequence fragment from one location to another
these movements are usually catalyzed by special enzymes which can involve insertions, deletions, and duplications depending on the mechanism of transposition
reversion is a process that restores a mutated sequence to its original sequence
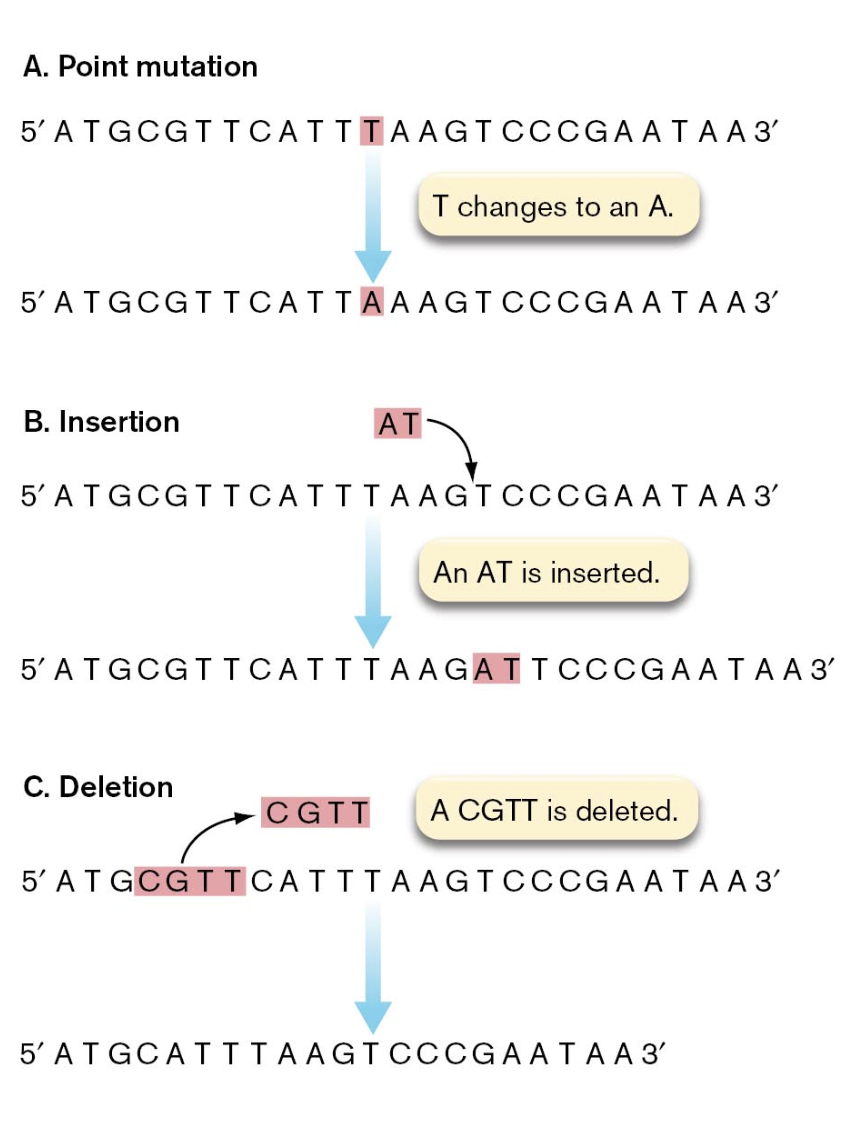
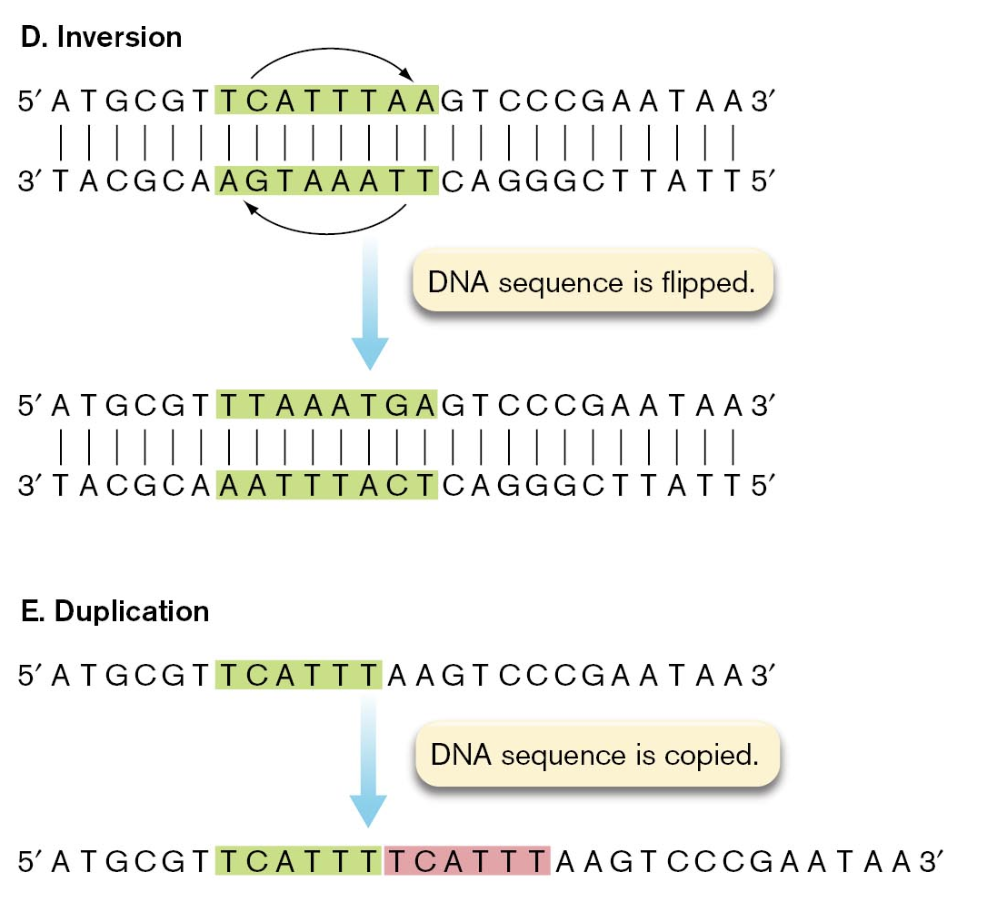
mutations can also be categorized into informational classes on the basis of how they affect the gene product
silent mutation — a mutation that does not change the amino acid sequence encoded by an open reading frame
the changed codon encodes the same amino acid as the original codon
missense mutation — a point of mutation that alters the sequence of a single codon, leading to a single amino acid substitution in a protein
there was a nonsynonymous base substitution
this sort of mutation may or may not lead to protein function alterations
the outcome will depend on the structural importance of the original amino acid and how close in structure & chemical properties of the replaced amino acid
missense mutations can result in either conservative amino acid replacements or nonconservative replacements
an example of a conservative replacement would be leucine substituted for isoleucine
an example of a nonconservative replacement would be tyrosine substituted for alanine
a missense mutation may decrease or eliminate the activity of the protein or it may make the protein more active / gain a new quality
loss-of-function — a mutation that eliminates the function of the gene product
gain-of- function — a mutation that enhances the activity or allows new activity of a gene product (eg expanded substrate specificity or gaining a completely different substrate specificity
INSERT IMAGE HERE
knockout mutation — a mutation that completely eliminates the activity of a gene product
these mutations can include multiple-base insertions and deletions & nonsense mutations
nonsense mutation — a mutation that changes an amino acid codon into a premature stop codon
an example of this would be TCA (serine) to TGA which results in a truncated protein
these truncations can completely knock out the function of proteins and more so in cases where the mutation occurred early in the reading frame
the defective proteins are degraded by cellular proteases
these mutations can turn genes into pseudogenes which can be lost from the genome during evolution
frameshift mutation — a gene mutation involving the insertion or deletion of nucleotides that cause a shift in the codon reading frame
these mutations often cause the ribosome to encounter a premature stop codon
these mutations can turn genes into pseudogenes
if the insertion or deletion involves multiples of three bases, the reading frame is not changed, but one or more amino acids are added or removed
duplications within an open reading frame can create repeats of codons and increase the length of the protein product
this type of mutation can occur if the length of the duplicated segment is not divisible by three
inversion mutation — a mutation that flips a DNA sequence
the rotation would retain the 5 prime to 3 prime polarity in a new molecule
if the inversion occurred within a gene, it would likely change the codons in the area and alter the resulting protein
these mutations often involve large tracts of DNA encompassing several genes meaning that if an entire gene with its promotor inverts, the gene will likely remain functional & its encoded protein may very well still be made
these mutations occur within a genome as a result of recombination events between similar DNA sequences or as a consequence of mobile genetic elements jumping between areas of a genome
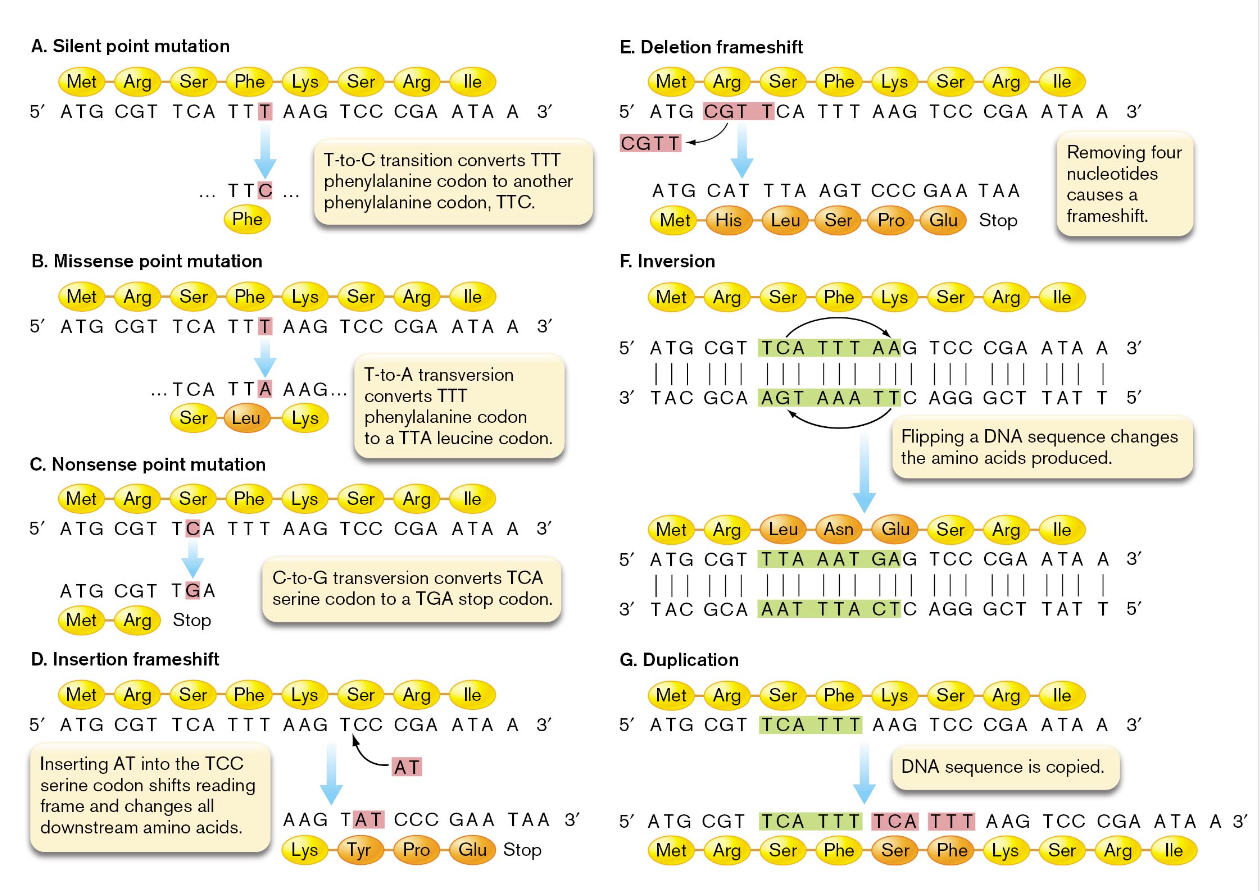
the effects that mutations have on the genome sequence can vary
mutations can affect both the genotype & the phenotype of an organism
every mutation causes a change in the genotype regardless of whether the mutation causes a change in the phenotype
recall that the phenotype only comprises of observable characteristics like biochemical, morphological, or growth traits expressed
the size of the mutation does not always correlate with the extent of a phenotypic change (usually dependent on the location instead)
mutations arise by diverse mechanisms
some mutations can arise spontaneously & despite the high accuracy of the replication machinery, mistakes do occur (at a low rate)
spontaneous mutations in a genome arise for many reasons
an example is tautomeric shifts in the chemical structure of the bases; these shifts involve a change in the bonding properties of amino and keto groups
these group forms usually predominate but when an amino acid shifts to an imino group, the base pairing changes
tautomeric shifts that occur during DNA replication will increase the number of mutation events
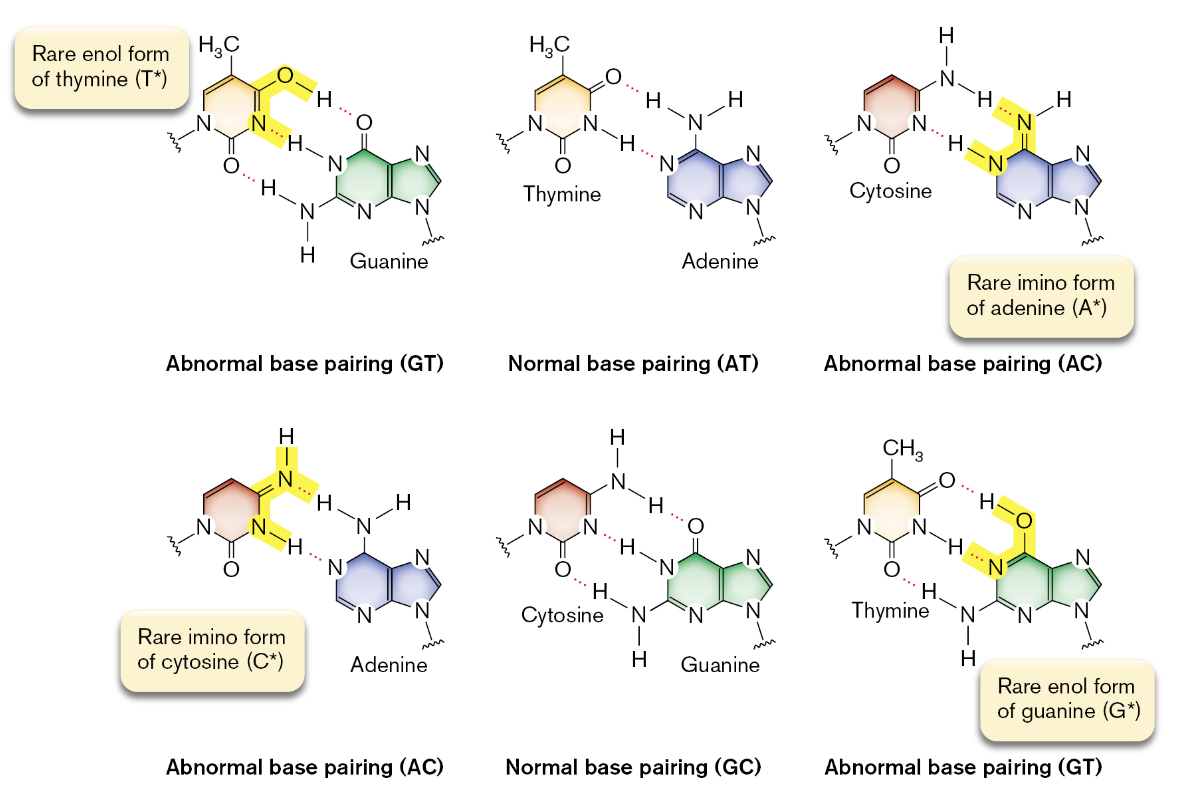
an example provided of how potentially detrimental tautomeric shifts can be is thymine shifting during replication which results in an AT to GC transition mutation
after the second round of replication, the mutation is “fixed” on both DNA strands in the mutant
the mutation will be passed onto all the mutant’s progeny
spontaneous mutations in DNA can also be caused by chemical reaction with water (hydrolysis)
an example of this is cytosine spontaneously deaminates to yield uracil which base-pairs with adenine instead of guanine
INSERT IMAGE HERE