Topic 2: Bacterial Cell Structure and Function
Prokaryotes vs Eukaryotes
Eukaryotes: cells that contain membrane bound organelles
ex. yeast, helminths, protozoa
Prokaryotes: cells that lack a membrane bound nucleus and other organelles
Main type we will study → bacteria
Viruses
neither eukarotic or prokaryotic
obligate intracellular pathogens - viruses cannot reproduce or carry out metabolic processes outside a host cell
Eukaryotic DNA
linear
double stranded
housed within the nucleus
packages - exists as a supercoil
diploid - 2 copies of every chromosome ( 1 maternal, 1 paternal)
mitosis (replication of somatic cells) vs meiosis (replication of gametes)
prokaryotic DNA
circular - single molecule
double stranded
located in the nucleoid region ( a visible mass)
nucleoid not seperate or protected in cell - just where DNA happens to localize
haploid → asexual, binary fission
plasmid (smaller, circular DNA molecules that exist separately from chromosomal DNA)
carry non-essential gene products
antibacterial resistance can be plasmid driven
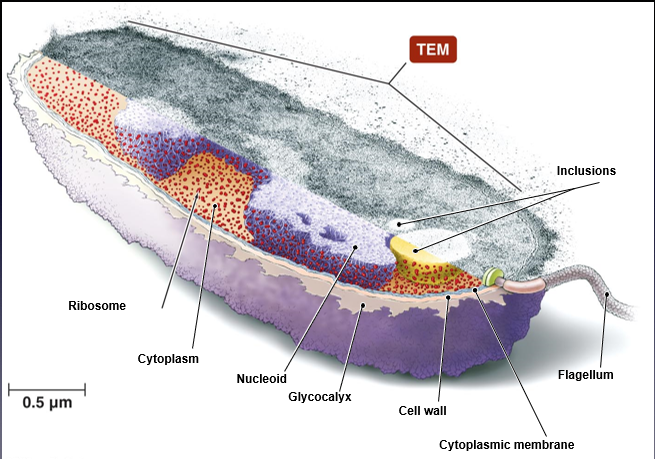
ribisome not membrane-bound → made of rRNA and protein
flagella are common in prokaryotes
THE BACTERIAL CELL
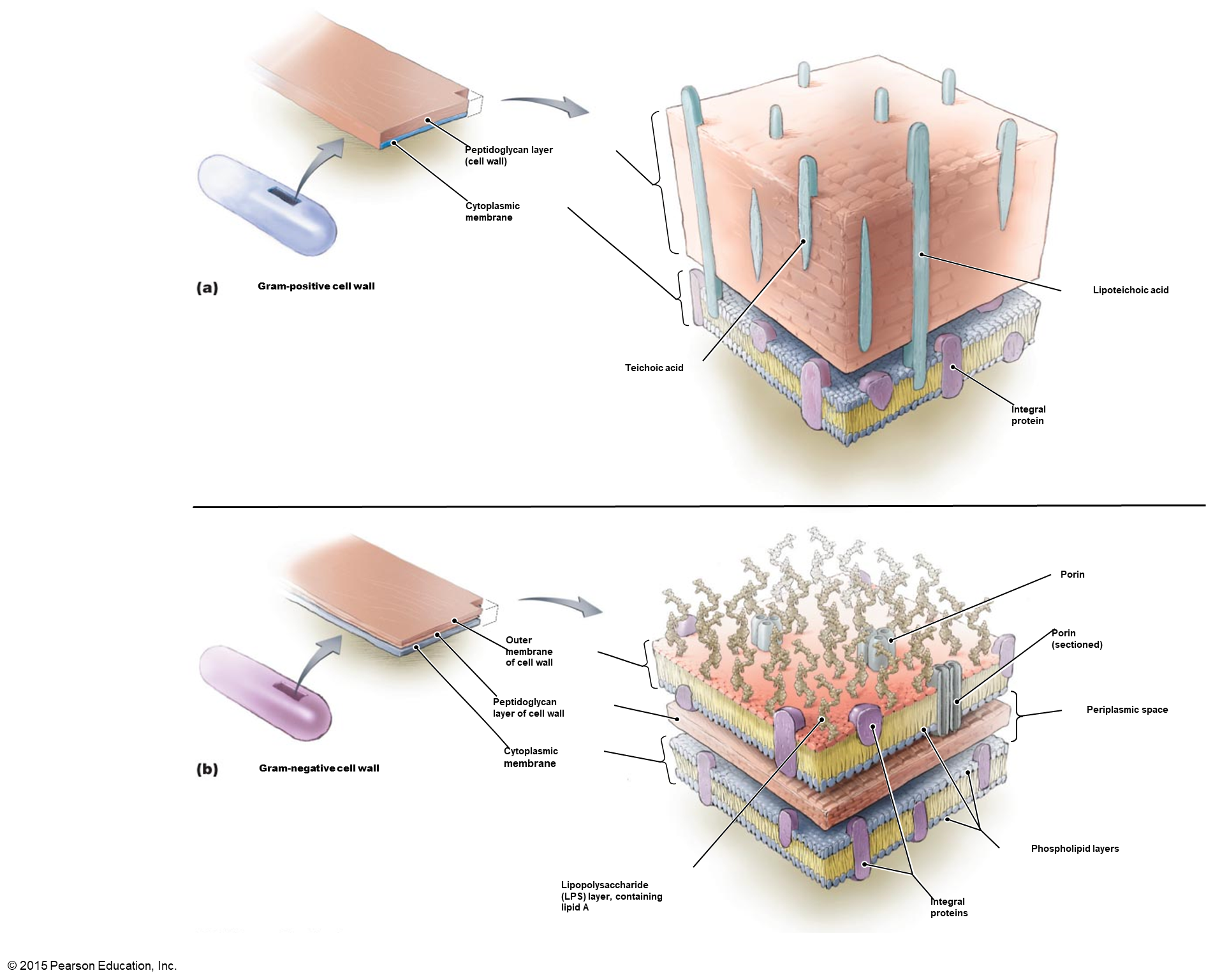
gram positive
thicker cell wall; more likely to be environmentally stable
ex. Clostridium tetani, Streptococcus, Staphylococcus, Bacillus anthracis
gram negative
thin cell wall; need host protection
ex. Yersinia pestis, Treponema pallidum, Helicobacter pylori, Bordatella pertussis
THE CELL WALL
G+ = thick (multiple layers) cell wall; G- = thin (single layer) cell wall
cell wall - complex semi-rigid structure
provides shape
external to the plasma membrane
provides protection
composition: peptidoglycan (nonhuman product)
Repeating disaccharides attached to chains of 4 amino acids
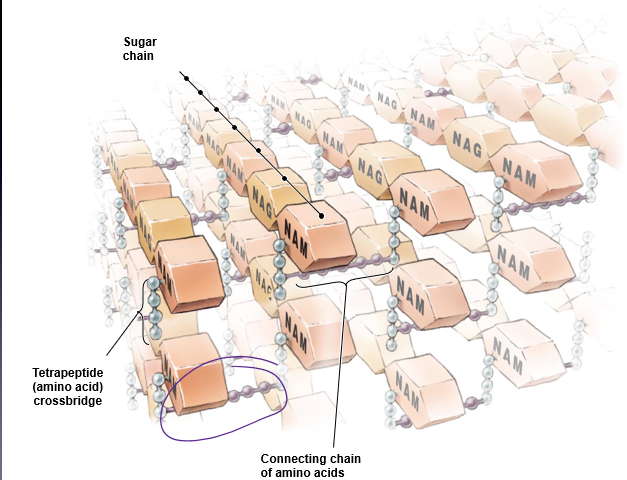
PEPTIDOGLYCAN STRUCTURE
disaccharide - two monosaccharides that alternate to form the carbohydrate backbone
N-acetyl-glucosamine (NAG)
N-acetyl-muramic acid (NAM)
Tetrapeptide chain (4 amino acids) attached to each NAM
Amino acids alternate stereoisomers of each other
Parallel tetrapeptide chains are generally linked by peptide cross bridges (1 to 5 amino acids) - hold peptidoglycan together!
*variation in length of carbon backbone → size of bacteria
**known target for medication → to stop bacteria from dividing

LPS will make humans incredibly sick
pepdydoglycan in between two membranes in G-
GRAM POSITIVE CELL WALL
up to 25 layers
type and strain variatin
peptide cross bridges will vary
5 L-Glycine in Staphylococcus
GRAM NEGATIVE CELL WALL
no characteristic peptide cross bridge
usually a peptide bond exists between DAP (#3) and Ala (#4) on different tetrapeptide side chains
PERIPLASMIC SPACE ***only in gram negative bacteria
space between outer surface of plasma membrane and the inner surface of outer membrane
12-15 nm thick
peptidoglycan found here
gel like consistency due to proteins
hydrolytic enzymes
binding proteins
chemoreceptors
PLASMA MEMBRANE
Encloses the cytoplasm
phospholipids & proteins - fluid membrane (bacteria have no cholesterol)
2 regions: Hydrophilic, Hydrophobic
Selective Barrier
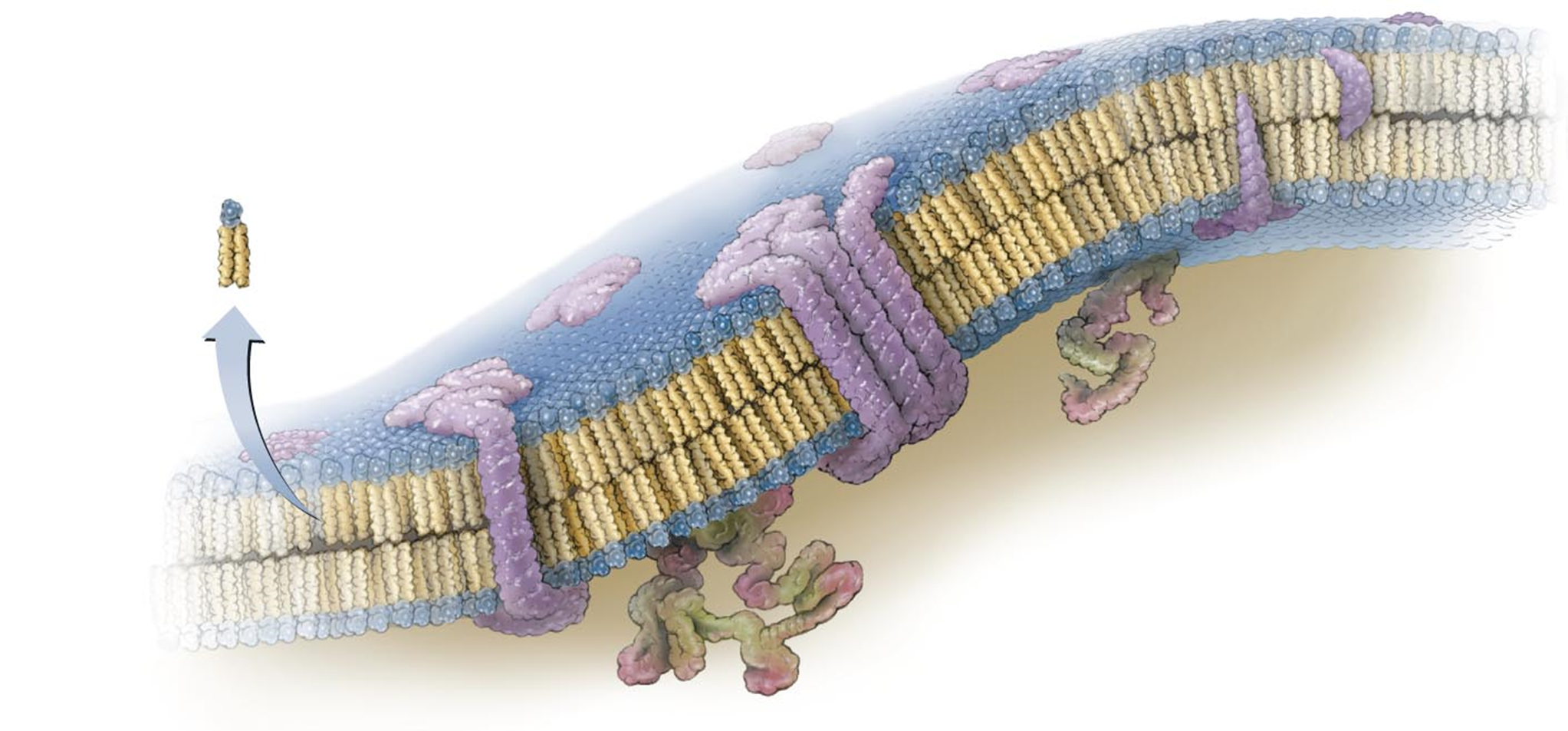
integral proteins span the entire membrane → ex. channels (purple)
peripheral proteins are more associated with one side
SELECTIVE PERMEABILITY OF PLASMA MEMBRANE
small hydrophobic molecules can diffuse through the membrane freely
3 systems of membrane transport for hydrophillin and charged molecules
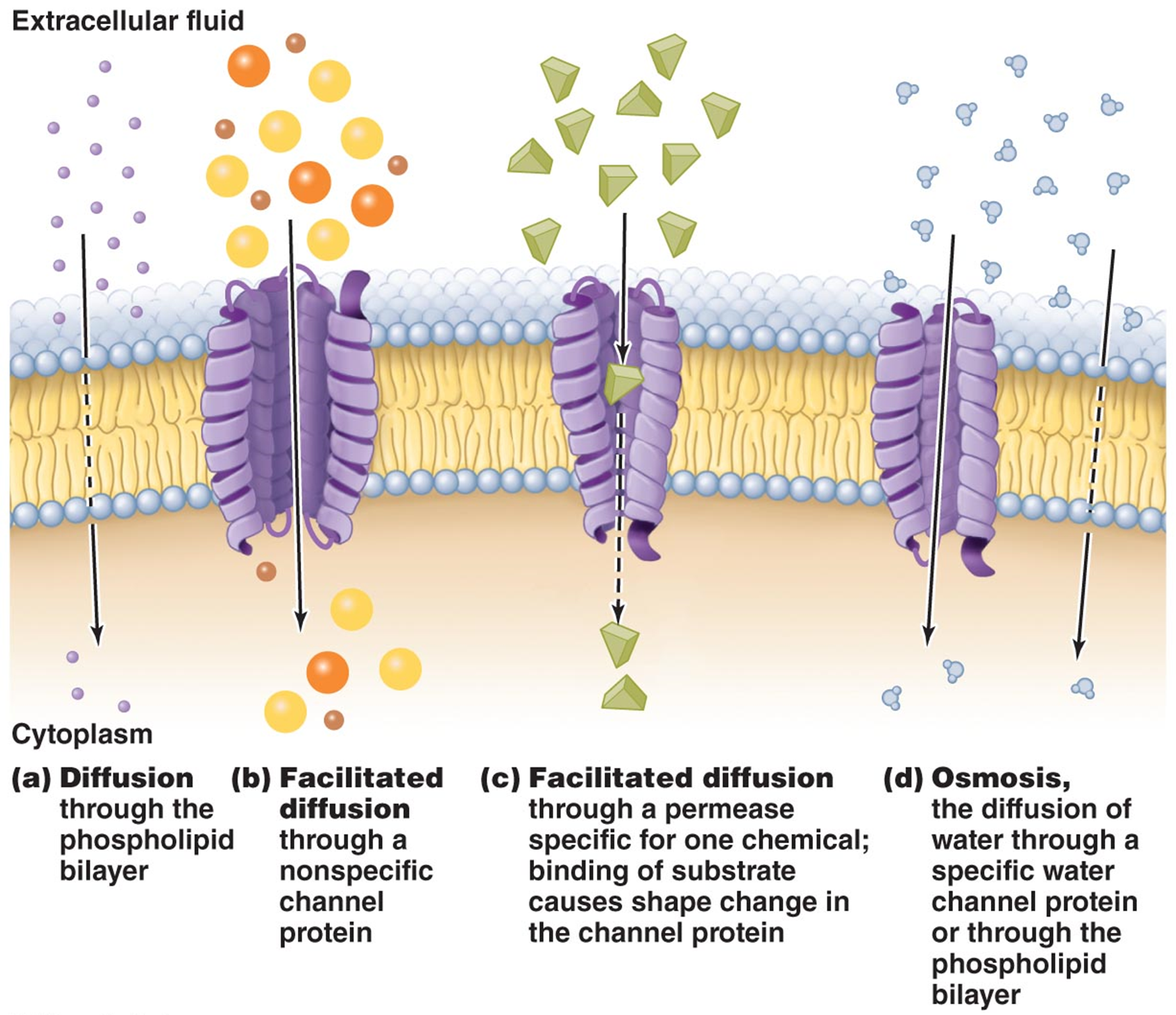
simple transport (w membrane spanning proteins)
uniporters, antiporters, symporters
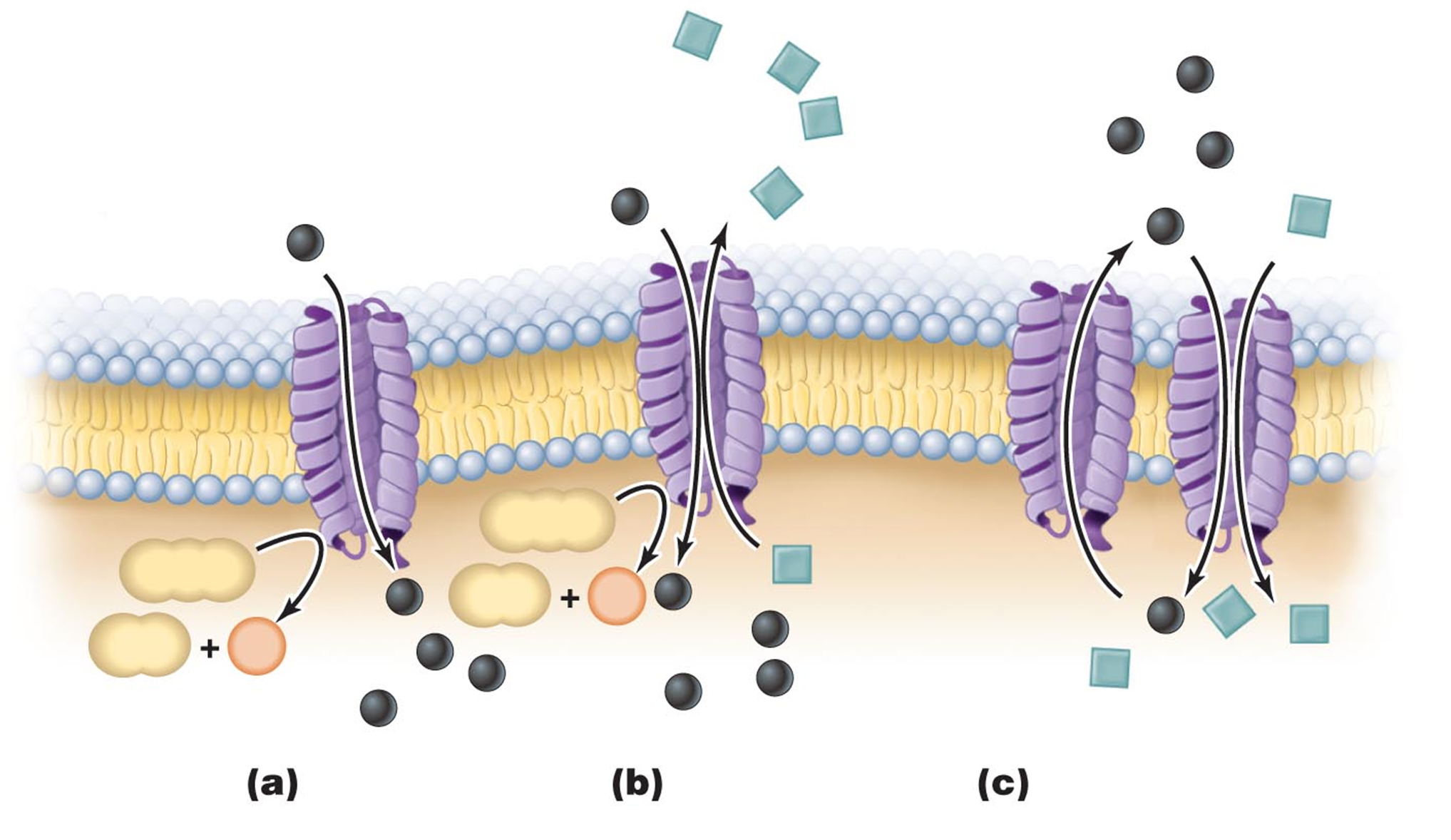
a. uniport b. antiport c. couples transport: uniport ad symport
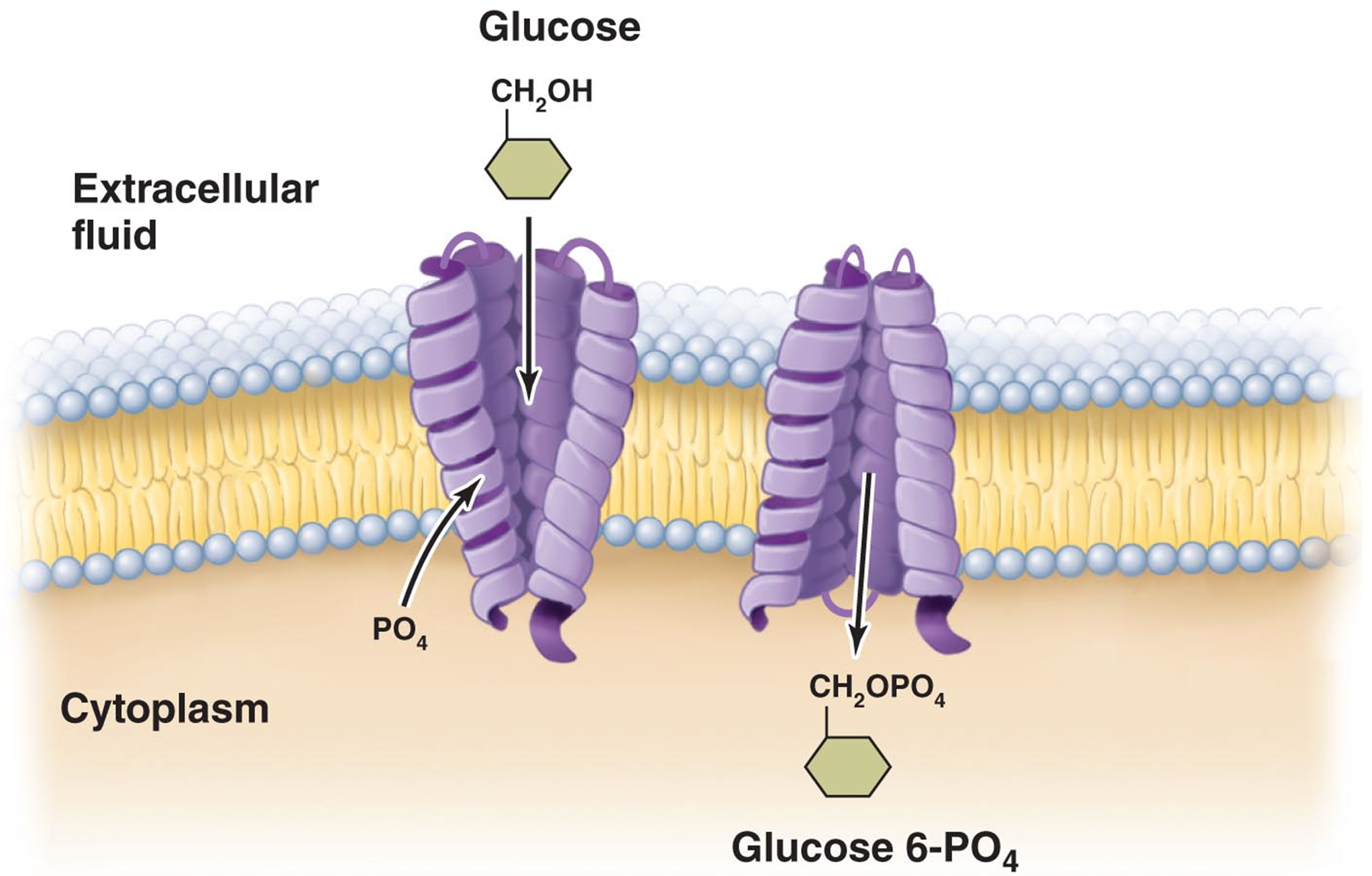
group translocation - substance is chemically altered during the transport process
ex. phosphotransferase system - transports glucose
system consists of 5 proteins - enzyme II C is the only one that spans the membrane
energy for this comes from phosphoenol pyruvate (glycolysis)
ABC system (ATP Binding Casette) - only in Gram negative
substrate binds to the periplasmic binding protein.
This binding triggers a conformational change, allowing the complex to interact with the membrane-spanning transporter.
The transporter then changes shape, driven by the energy released from ATP hydrolysis, allowing the substrate to be transferred into the cell.
** similar transport mechanism exists in Gram + bacteria.
the initial binding proteins are anchored directly in the plasma membrane rather than being located in the periplasmic space.
OUTER MEMBRANE of G-
consists of bilayer of phospholipids, imbedded proteins, Lipopolysaccharide
all contribute to prevention of perplasmic enzymes diffusing away
porins are specialized channels that allow passage of small molecules and ions
LPS tend to make human very sick
LPS components
O polysaccharide - repeating unit of 4-5 sugar residues
varies between differents strains and species of bacteria
core polysaccharide - connects O poly to Lipid A
includes ketodeoxyoctanate (KDO), a sugar that links to other sugars
Lipi A - achors the entire structure to the outer membrane
“endotoxin” - released when cell is lysed during infection
induces fever, blood clotting (decreased BP)
decreased BP can cause organ failure, shock, death
**positive feedback loop
CYTOPLASMIC INCLUSIONS
reserve deposits (prokaryotes do not have vesicles)
2 main types
PHB (poly-β-hydroxybutryric acid) - Lipid like
Carbon and energy storage
Polyphosphate granules
Inorganic phosphate - DNA/RNA synthesis
The glycocalyx (sugar structure) is a sticky layer composed of polysaccharides, polypeptides, or both. It can take two primary forms: capsules and slime layers.
CAPSULES AND SLIME LAYER
found outside of the cell wall in some bacteria
Capsule - typically thick; firmly attached to cell wall
Slime Layer - typically thin; loosely attached to cell wall
Sugar “shell” protects from immune system detection
Cells in immune system is looking for proteins – so immune system unable to detect bc these shells cover proteins
what do they do??
Capsules - contribute to bacterial virulence
Protect disease causing bacteria from phagocytosis by leukocytes
Capsules & Slime Layers - attachment to surfaces
S. aureus (capsule) and S. epidermidis (slime layer) - high affinity for titanium
Titanium in joint replacements → staff can make its way into the bone cells and live inside the bones → antibiotics cant penetrate solid matrix so must cut the bone to reach
FLAGELLA
long filamentous appendages
G+ or G- motile bacteria - propel organism
3 distinct arrangements

completely covered by flagella
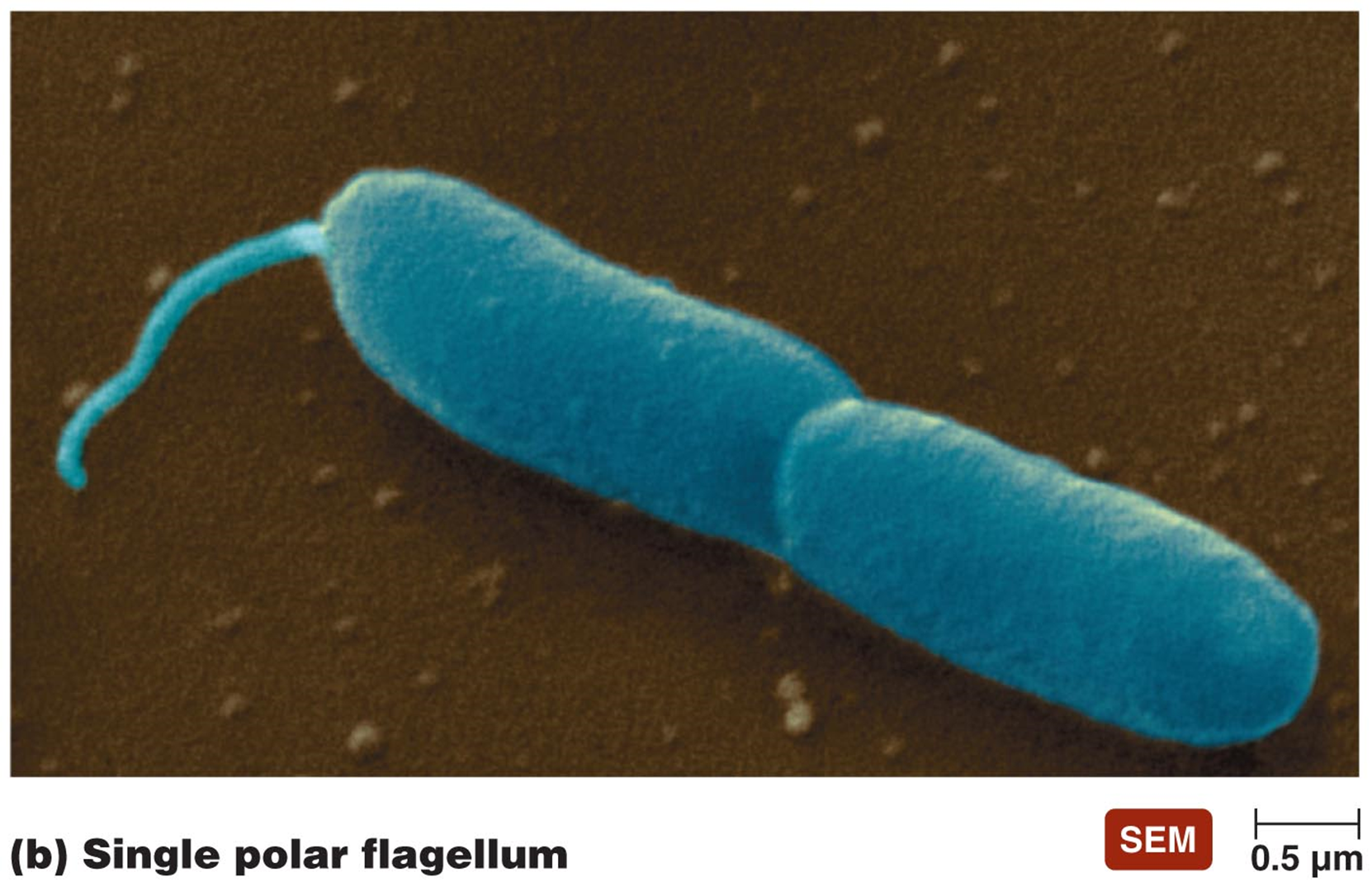
polar
could have one at each end
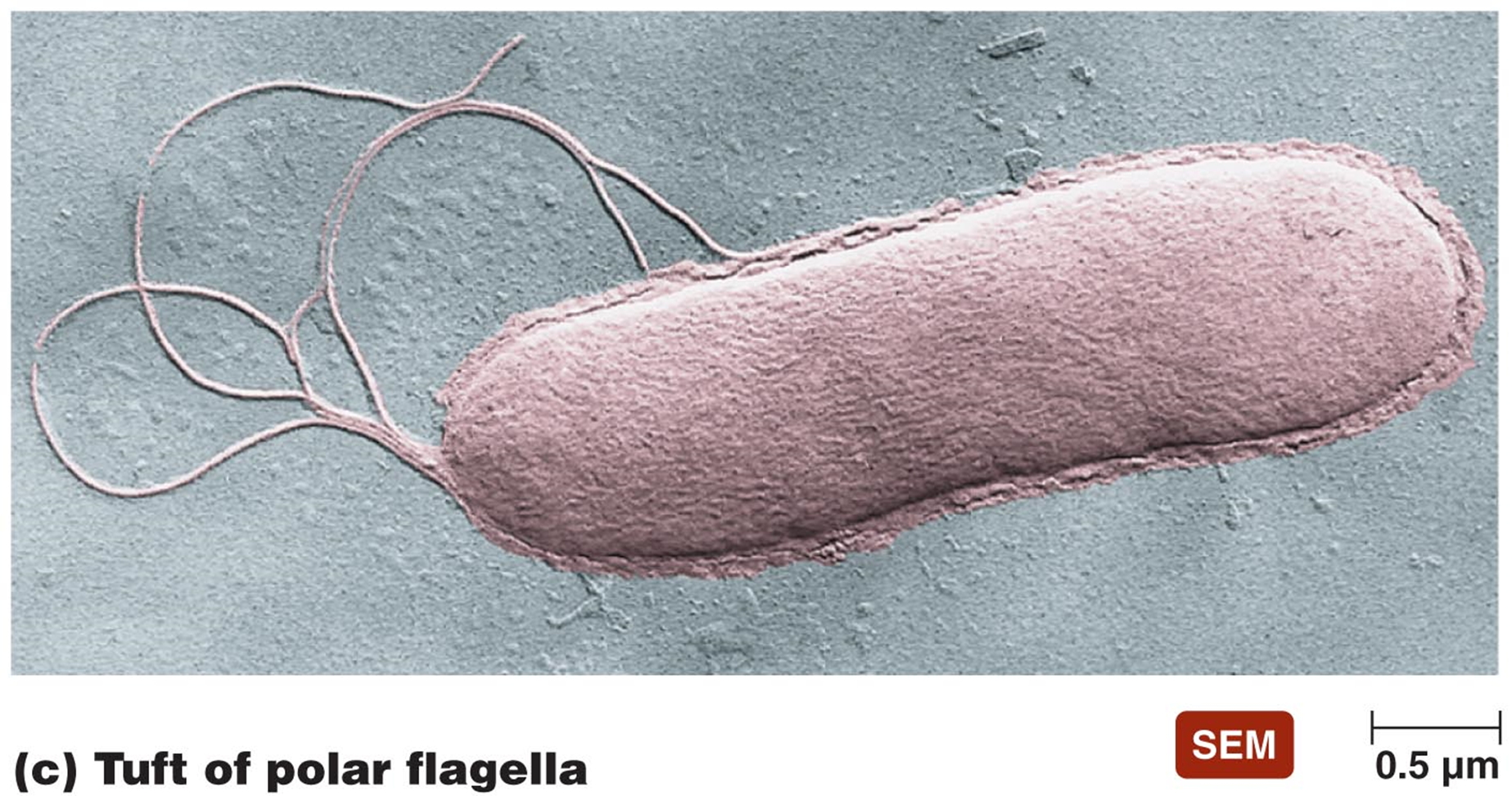
lophotrichous
multiple coming from one or both ends
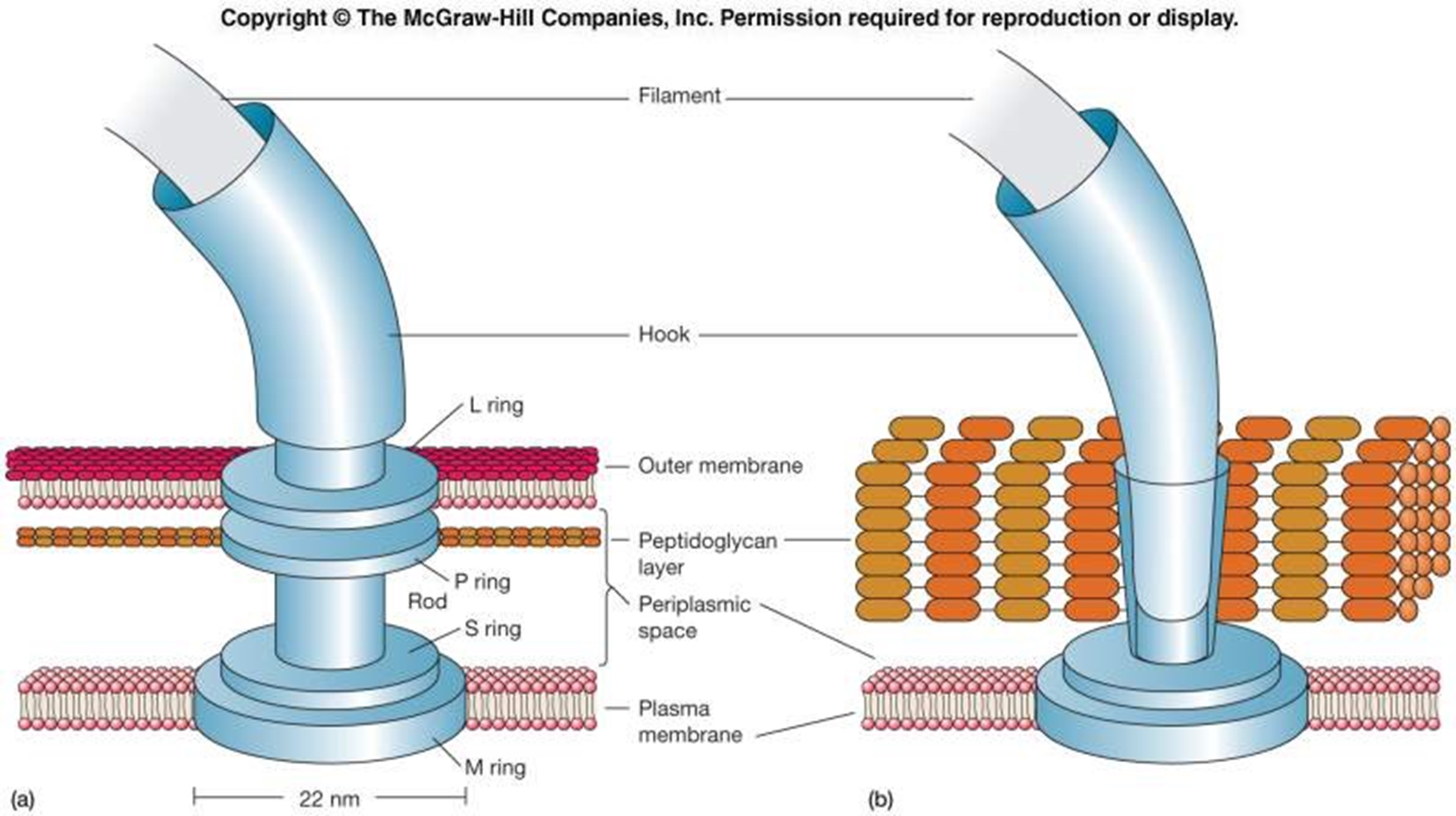
FLAGELLAR STRUCTURE
Filament - long, outermost region with subunits of flagellin protein
Several chains intertwined around a hollow core
Hook - wider region at base
Attaches the filament to the motor
Motor - a biological motor - central rod through a system of rings (L ring, P ring, MS ring, C ring)
Embedded in outer membrane (L ring), peptidoglycan (P ring), plasma membrane (MS ring) & cytoplasm (C ring)
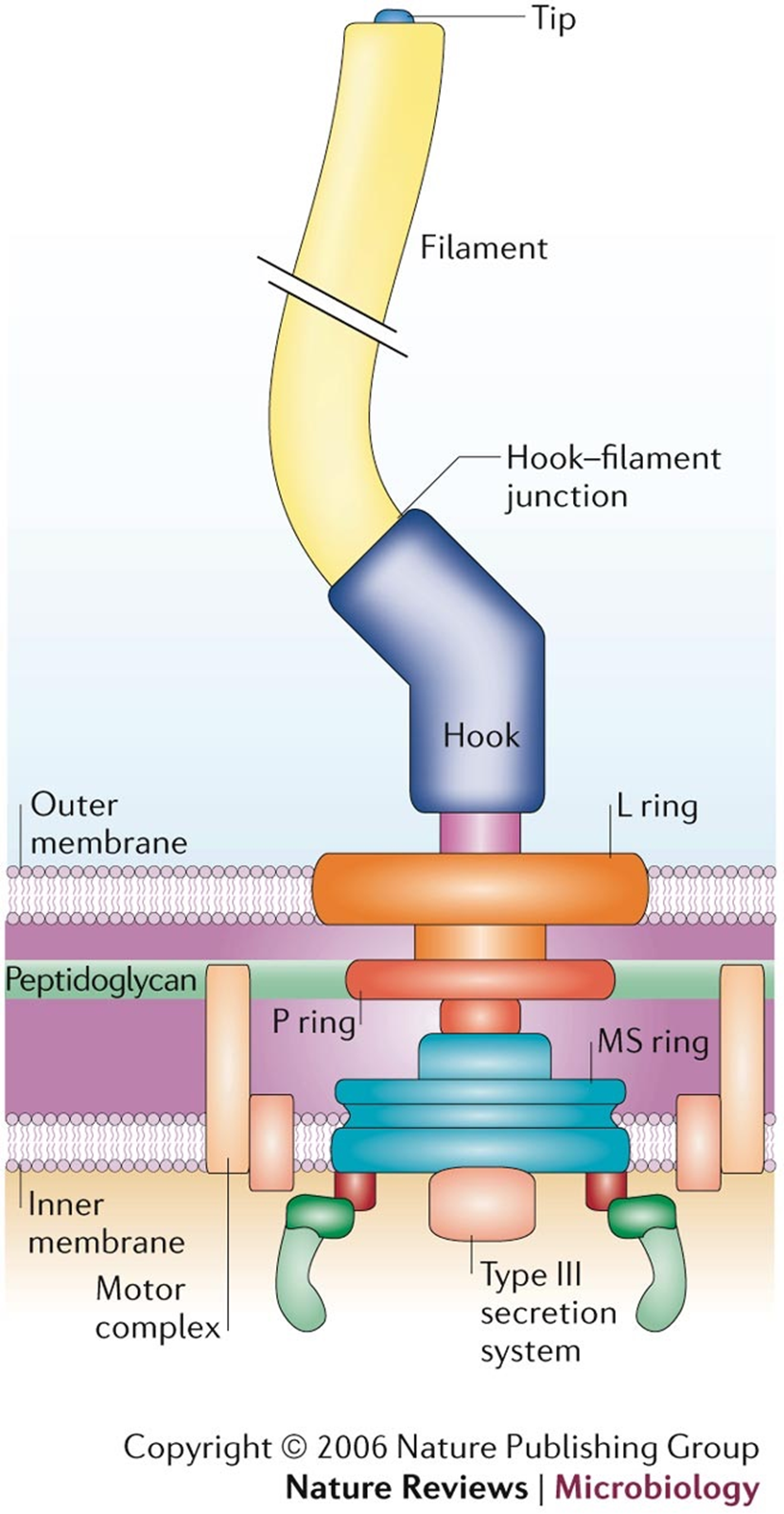
All 4 rings found in G - (bc it has an outer membrane)
** so not all rings in G+ ???
Flagellin is prokaryotic; tubulin is eukaroytic
**Immune response from receptors that detect flagelli → not a natural human protein
EUKARYOTIC FLAGELLA
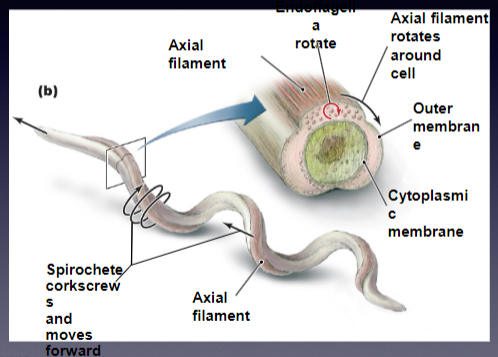
AXIAL FILAMENTS
consist of flagella that are located within the periplasmic space
could run the entire length of the cell
**spirochete allows for corkscrew motion (of the entire bacterium)
BIOLOGICAL MOTOR
the flagellar motor consists of mot and fli proteins → essential for both the motor function and the switching mechanism that controls direction.
proton motor force - As protons flow down their concentration gradient through the motor proteins, they provide the energy needed for the motor to function.
When protons(+) move through rings (+), they create a repulsive force → leads to the rotation of the motor, which in turn causes the flagellum to spin.
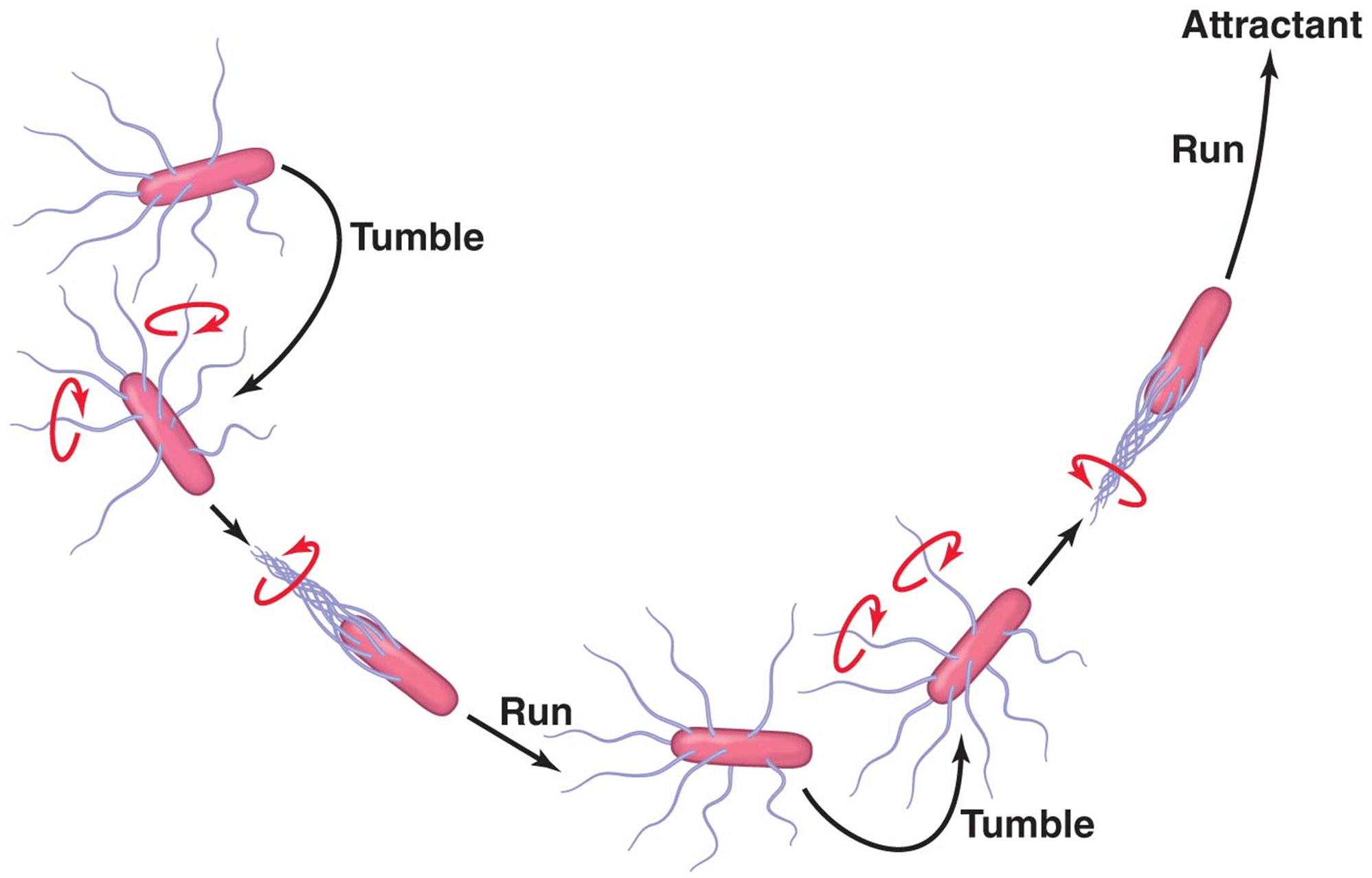
CW motion - tumbles (random series of movement)
CCW - runs (coordinated movement toward stimulus)
CELL TAXIS
Cells may move towards or away from a stimulus
Favorable stimulus - increased runs
Unfavorable stimulus - increased tumbles
Chemotaxis & Phototaxis
Chemo/photoreceptors detect stimuli - interact with other proteins to affect flagellar motion
PILI
Hair like appendages on some bacterial cells → Shorter and thinner than flagella
helical structure of protein pilin chains around a central core
With few exceptions, generally G- only
functions
Attachment - facilitate attachment to surfaces, including host tissues.
ex. gonorhea attaches to mucus - changes expression on surface of cell
Conjugation
Certain pili, known as sex pili, are involved in conjugation, a form of horizontal gene transfer.
donor bacterium forms a conjugation bridge through its pili to connect with a recipient bacterium, facilitating the transfer of genetic material (such as plasmids). This is essential for the spread of antibiotic resistance and other traits among bacterial populations.
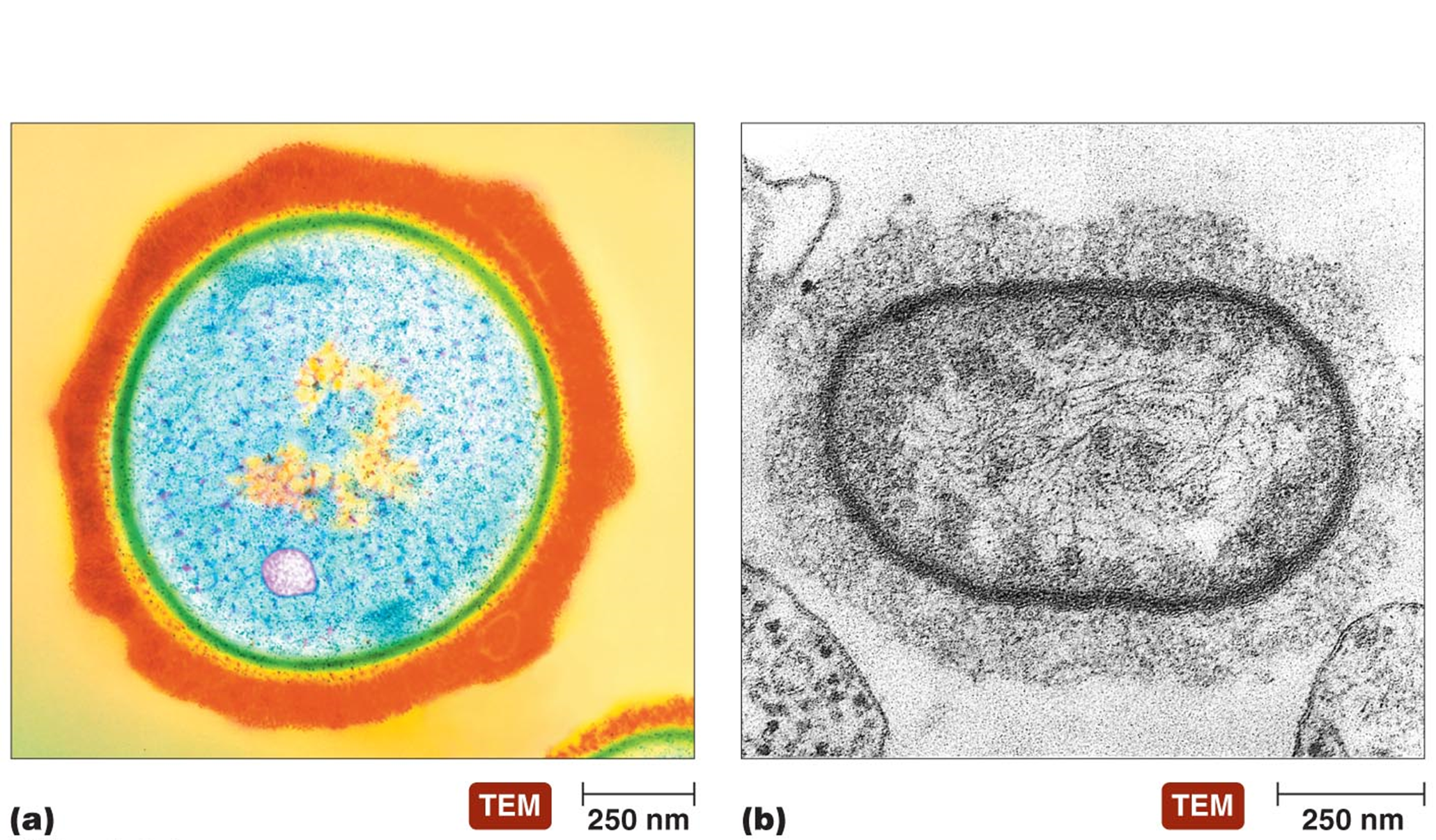
ENDOSPORES (structures formed by certain bacteria)
found primarily in G+and very few G-
2 genera that cause human disease - Clostridium & Bacillus
Highly durable bodies with thick walls
Formed when essential nutrients or water is lacking (not ideal circumstances)
Not reproductive
Survive extremes and germinate when conditions are favorable (could be an animal cell) → tranforms to the vegetative (active) cell
can remain dormant for many years
**Anthrax Island

endospores can survive extreme heat, lack of water, toxic chemicals, radiation
**can be elimated with extreme heat under pressure (autoclave), chrorine dioxide gas
KEY COMPONENTS OF ENDOSPORES
Dipicolinic Acid - characteristic of endospores
Found in core of endospores
Complexes with Ca2+ to give gel like structure → decreases water content
Small Acid Soluble Spore Proteins (SASPs) - bind tightly to DNA in core, protect DNA from desiccation, radiation and heat
also serves as a Carbon & energy source when endospore germinates and transitions to vegetative cell
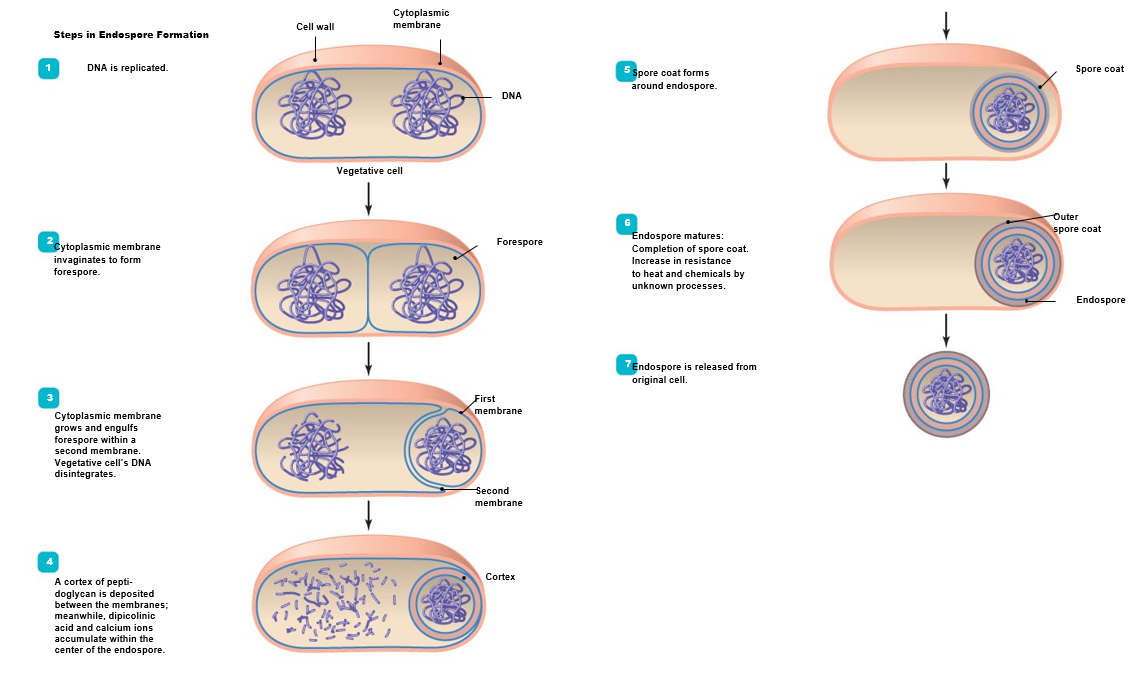
ENDOSPORE FORMATION (6-8 hours) (in stressful environment)
DNA replicated
Forespore formed
Plasma membrane engulfs forespore
Cortex of peptidoglycan formed
Spore Coat formed
Endospore released from vegetative cell