Gene Control
Transcriptional Regulation
Operator: sequence of DNA to which the transcription factor protein binds
Promotor: sequence of DNA to which RNA polymerase binds to start transcription
General Transcription Factors (GTFs): a class of proteins that bind to specific DNA sites to activate transcription
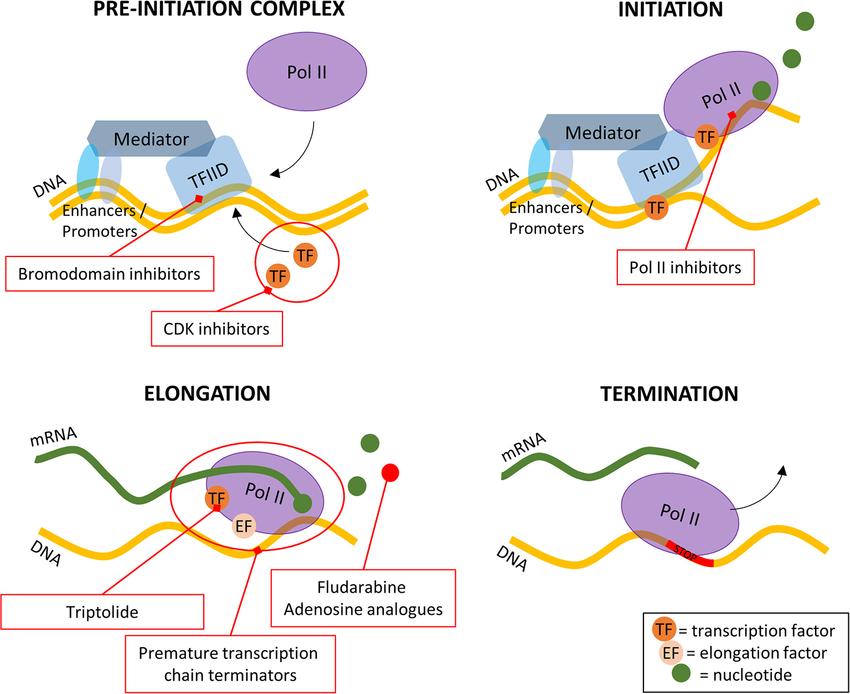
The basic transcription apparatus consists of..
GTFs+ RNA polymerase +mediator multiple protein complex
the apparatus positions RNA pol right at the start of the protein coding sequence (gene) and then releases RNA pol to transcribe mRNA from the DNA template
Activators (another type of DNA binding protein): enhances interactions between RNA pol and a particular promoter, encouraging gene transcription.
activators do this by increasing the attraction of RNA pol for the promoter through either interactions with the RNA pol subunits or indirectly by changing the structure of DNA.
ex: catabolite activator protein (CAP) activates transcription of the lac operon in E. coli
CAmp ( catabolite activator protein) is produced during glucose starvation, binds to CAP and causes conformation change which allows the CAP to bind to DNA site adjacent to the promotor. CAP then makes direct protein-protein interactions that recruits RNA pol to the promotor.
Enhancers: sites on DNA bound to by activators in order to loop the DNA in a certain that brings a specific promotor to the inhiation complexes.
AKA they enhance transcription of genes in a particular gene cluster
they are usually cis-acting (they act on the same chromosome), but they don’t need to be particularly close to the gene they’re acting on
enhancers don’t act on promotor region themselves, but are bound by activator proteins which can interact with mediator multiple protein complex (complex recruits RNA pol, and GTFs leading to the transcription of the gene)
Repressors: proteins that bind to the operator, impeding RNA pol. movement along the strand and thus impeding transcription/expression of the gene.
if an inducer ( molecule that initiates gene expression, like lactose with the lac operon) is present, it can interact with repressor in a way that causes it to detach from the operator so RNA pol. is free to further transcribe the gene.
Silencers: regions of DNA bound by repressor proteins in order to silence gene expression
similar mechanism to that of enhancer sequences and can be located several bases upstream or downstream from the actual protein.
when a repressor protein binds to silencer region, RNA pol. is prevented from binding to the promotor
![]()
Differences between prokaryotes and eukaryotes in transcriptional regulation
Prokaryotes: transcription regulation is needed for the cell to quickly to adapt to the everchanging environment the cell is sitting in.
presence, quantity, and type of nutrients available determines what genes are expressed
Prokaryote regulation is mostly activators and repressors, rarely enhancers
Eukaryotes: transcriptional regulation tends to involve a combination of interactions between several transcription factors, allows for more sophisticated responses to the environment
eukaryotes also have a nuclear envelope, which prevents simultaneous transcription and translation, allowing an extra spatial and temporal control of gene expression.
Post-Transcriptional Regulation (occurs in Eukaryotes)
DNA gets transcribed base-for base into pre-mRNA. Then this pre-mRNA strand needs to be modified before it leaves the nucleus as fully processed mRNA. Processing helps stabilize/protect the mRNA.
ONLY EXONS MAKE IT INTO FINISHED mRNA. they code for the ultimate protein product.
Introns: short non-coding segments of RNA that get cut out of spliced out.
RNA splicing is done by a spliceosome, a large molecular entity (a snRP) that binds on either side of the intron, loops it into a circle and then cleaves it off and ligates the two ends of exposed strands together.
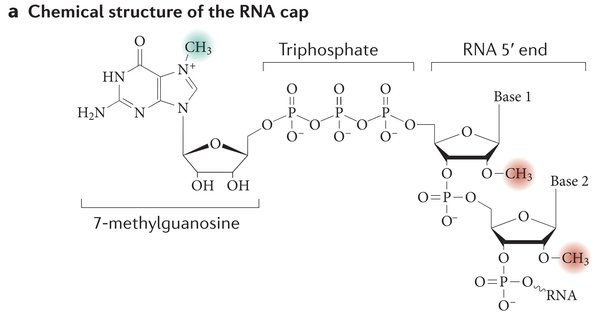
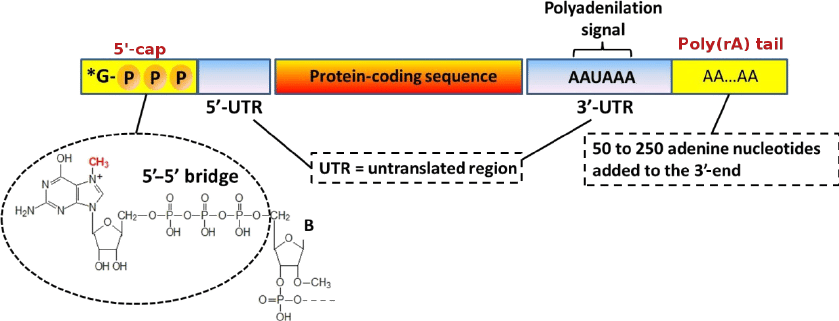
After splicing, the RNA receives a 5’ cap and a 3’ poly-A tail, which helps stabilize the mRNA for translation.
the 5’ cap is on the phosphate end (five=fosphate). The cap (a guanine nucleotide) coverts that end of the mRNA to a 3’ and by a 5’-5’ linkage (the monophosphate of the RNA becomes triphosphate by precursor reactions for the addition of the cap).
the cap protects the mRNA strand from exonucleases, promotes ribosome binding, and regulates nuclear export of the mRNA.
The Poly-A tail goes on the 3’ end. Multiple adenosine monophosphates are added to act as a buffer for exonucleases. This increases half life of mRNA and again protects from degradation, promotes translation by ribosomes, and regulates nuclear export.
the poly-A tail also helps with transcription termination for RNA polymerase
poly-adenylation is catalyzed by the enzyme polyadenylate polymerase, which adds adenine molecules using ATP as a substrate. The poly-A tail is built until it’s about 250 nucleotides long.
 RNA Editing: results in sequence variation in mRNA molecule. Relatively rare but has many catalysts.
RNA Editing: results in sequence variation in mRNA molecule. Relatively rare but has many catalysts.
may include insertion, deletion, and/or substitution of nucleotide bases on mRNA molecule
“Adenosine Deaminase Acting on RNA” aka ADAR is one type of RNA editing which converts specific adenosine residues to inosine in an mRNA molecule by hydrolytic deamination.
CDAR: involves deamination of cytosine to uridine by cytosine deaminase
RNA editing is studied for infectious diseases because of editing process alters viral enzymes
Non-Coding RNA (ncRNA)
ncRNA is a functional RNA molecule that is not translated into a protein. It performs vital functions in the cell still as RNA. (most participate in transcription and translation in one capacity or another)
MicroRNA (miRNA): functions as transcriptional and post-transcriptional regulation of gene expression by base paring with complementary sequences within mRNA molecules.
Ribosomal RNA (rRNA): helps make up ribosomes, used in translation
Transfer RNAs (tRNAs): links codons in mRNA strand to corresponding amino acid for polypeptides
Small nucleolar RNA (snoRNA): class of small RNA molecules that guide covalent modifications of rRNA, tRNA, and snRNA through methylation or pseudouridylation (addition of an isomer of nucleotide uridine). Also relevant during translation.
Small nuclear RNA (snRNA): avg length is 150 nucleotides. Primary function is the processing of pre-mRNA in the nucleus. They also aid in the regulation of transcription factors or RNA polymerase II, and maintaining telomeres.
can be associate with a specific set of proteins that form complexes called snRNPs
Spliceosome (snRNA + snRNP) which removes introns during processing of pre-mRNA. Performs binding of introns and two sequential trans-esterification reactions that splice out introns and ligate exons to form mature mRNA