DNA Structure and Analysis
Vocabulary:
Genetic material: Responsible for inheritance, controls the form and characteristics of individuals, includes forms of DNA and RNA , the genetic material of all living organisms. Must be able to replicate, store and express information, and allow variation by mutation
Replication: The process that copies genetic material, ensures that new cells receive the same copy of heritable material. Essential to growth/renewal of cells, a fundamental property of all living things
Information storage: A property of genetic material, they encode all the information needed to transmit traits to progeny
Information expression: A property of genetic material, despite having the same base genetic material, the way it is utilized varies, allowing only specific parts of an organism to acquire certain form and perform unique actions
Variation: Occurs via mutation to genetic material, leads to inherited changes that can be distributed amongst the population, the raw material process of evolution
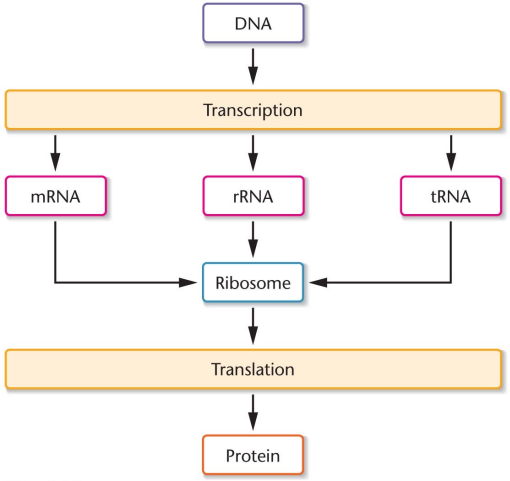
Central Dogma of molecular genetics: Information in DNA is transcribed into the chemical language of RNA, and then translated into genetic information or amino acids, only flowing in one order
Friedrich Miescher: Swiss chemist and biologist, discovered DNA by studying white blood cells in pus and extracting the nuclei, they were rich in phosphorus and called in nuclein, one of his students called them nucleic acids
Frederick Griffith: Conducted experiments in 1928 on streptococcus pneumonia and found that heat killed bacteria, and he could turn non-pathogenic bacteria into pathogenic ones. Showed the capsule alone wasn’t the cause of death, speculation about the transforming principle. S (smooth, had a polysaccharide coat) strands were pathogenic, R (rough) weren’t. In his experiment, he put bacteria in a mouse that didn’t have the capsule (R) and then found the mice dead with capsules (S) so there was a transforming factor passed on
Transformation: Transfer that bacteria undergo
Avery, MacLeod, and McCarty: In 1944 did many experiments on purified fractions of the transforming factor and concluded that genetic material was actually DNA, as it was the fundamental unit of the transforming principle in pneumococcus type III
Alfred Hershey, Martha Chase: In 1952 showed DNA is the genetic material of the T2 phage, as only DNA enters E. Coli cells during infection. Concluded that the capsule of a virus remains on the outside of a cell, that is made of protein and the DNA is injected into the cell. Sulfur is only in proteins, phosphorus is only in DNA, using radioactive isotopes and a centrifuge found sulfur only in shells, but phosphorus made more phages with more radioactive phosphorus
UV Light: Can cause mutations in genetic material, most mutagenic at 260 nm, where it is more strongly absorbed by DNA/RNA than proteins
Recombinant DNA: Direct evidence that DNA is genetic material, genes can be taken from humans and put into plasmid vectors which go into bacteria cells, which will then produce functional eukaryotic proteins
Primary structure: Linear order of nucleotides
Secondary structure: Double helical structure
Tertiary structure: Compacting/packing of DNA and protein in the nucleus around histones
Pentose sugar: Deoxyribose in DNA (2’ only has hydrogen) ribose in RNA (2’ has OH group)
Phosphate group: Negatively charged
Purine: Adenine or Guanine, a double ring
Pyrimidines: Cytosine, Thymine/Uracil, have a single ring
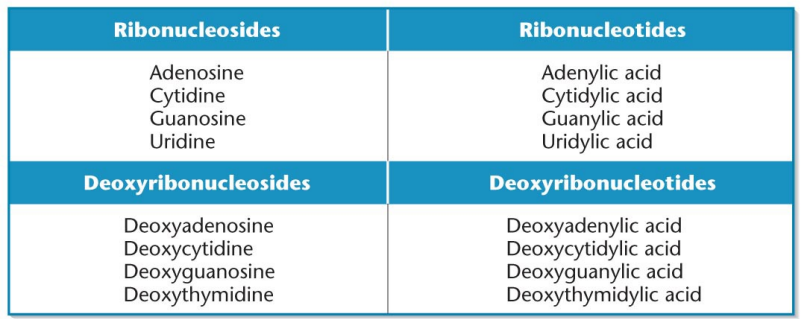
Nucleoside: has the nitrogenous base and pentose group, precursor to the nucleotide and other nucleic acid molecules
Nucleotide: A nucleoside with an attached phosphate group or multiple phosphates, attached to the 5’ carbon. Multiple is di and tri phosphate
Phosphodiester bond: Connects two nucleotides, covalent bond
Tetranucleotide theory: Incorrect, thought DNA was 4 base units without variation
Edwin Chargaff: Measured the amount of base pairs by paper chromatography to measure nucleotide ratios in various organisms, C matched G and A matched T. Made Chargaff’s rules
Chargaff’s rules: A/t equal, C/G equal, the base composition of DNA varies between species
Watson, Crick: Made a DNA model based off available data, stole Franklin’s work but still got the nobel prize. Introduced the idea of complementary base pairing and antiparallel strands
Rosalind Franklin: Got a picture of DNA via X-ray crystalography. Figured out nitrogenous bases paired and there were 2 sugar/phosphate backbones
Secondary DNA structure: A right handed double alpha-helix, nucleotides have nucleosides lying parellel while phosphate groups are vertical. Held together by hydrogen bonds. The diameter is 20A (angstroms) across
Major groove: Part of secondary DNA structure, space left by how DNA spirals unevenly, larger. This space allows for more accessablity for proteins like those for transcription, or anything sequence reliant. Factors for gene regulation, replication, and repair all bind here
Minor groove: Part of secondary DNA structure, space left by how DNA spirals unevenly, smaller. Specific non-regulating proteins access DNA here like DNA polymerase, ribosomes, and histones. Shape recognition comes from van der waals forces
B-DNA: A conformation of DNA that likely exists in aqueous cells, biologically significant, described by Watson-Crick model. One full turn every 10 base-pairs
A-DNA: More compact, exists under dehydrated or salty conditions (like in prokaryotes), used in protective scenarios in extreme conditions
Z-DNA (zig-zag): Forms a left-handed helix, longer and thinner than typical DNA, may contain large segments of C-G bases, can relieve torsion stress
BZ Junction: Places between B and Z DNA, might happen more with cancer mutations, have a extruded base pairs
Ribozyme: RNA that functions like certain enzymes due to its secondary shape, build from hairpins and stems
mRNA: Used in translation, template for protein synthesis, transferred to the site protein synthesis, short lived messengers
rRNA: Structural components of ribosomes, around 80% of all RNA
tRNA: Link between DNA code and amino acid order, carry amino acids for protein synthesis, between 75-90 nucleotides
Sedimentation behavior: How something will behave once in a centrifuge, depends on density, mass, and shape as well as viscosity and centrifugal force applied
Svedberg coefficient (S): A measure time affected by size, shape, and density
Retrovirus: Replicates unusually through reverse transcription, DNA intermediate is incorporated into the host’s genome, meaning when the host DNA is transcribed more retroviral chromosomes are produced
Reverse transcription: The process by which some RNA is the template for single stranded DNA synthesis, before the DNA is transcribed. Happens to retroviruses
Reverse transcriptase: RNA-dependent DNA polymerase, large factor in reverse transcription
Hyperchromic shift: The phenomenon where UV light is absorbed by dsDNA at an increasing rate as DNA is denatured. Purine/Pyrimidine more strongly absorb UV
Denaturation: The process of DNA breaking down, occurs at high heats. Hydrogen bonds break causing bases to separate through no covalent bonds break. Turns less viscous, and absorbs UV better
Melting point (Tm): The midpoint on a melting curve of temperature plotted against OD260, led to discovery that CG rich samples denature slower (contain one extra hydrogen bond). Also impacted by DNA length
Molecular rehybridization: The process of reversing denaturation, only of a reasonable degree of complementation exists. Done by cooling melted DNA slowly, allows slow reassociation
Reannealing: Part of molecular rehybridization, DNA and DNA strands reform
In situ hybridization: Part of molecular hybridization, when ssDNA or RNA probes are used to form complementary base pairs with DNA/RNA resent in tissue/chromosome sample, with fluorescence is called FISH
Electrophoresis: Technique used to separate charged molecules according to size. Particles move through agarose gel when exposed to electrical current. Ends of the chamber have different charges, and molecules are pulled towards the opposite ends and migrate. Since smaller molecules travel faster they travel further and can be separated by size
DNA Ladder: Molecular marker, quantifies DNA products. Solution of different known molecule lengths, used to reference accurate size of DNA fragments in gel electrophoresis
Originally proteins were thought to be genetic material as they were far mor complex than DNA, after experiments with phages it was known that at least prokaryotes used nucleic acids for their genetic material, would be known for eukaryotes much later
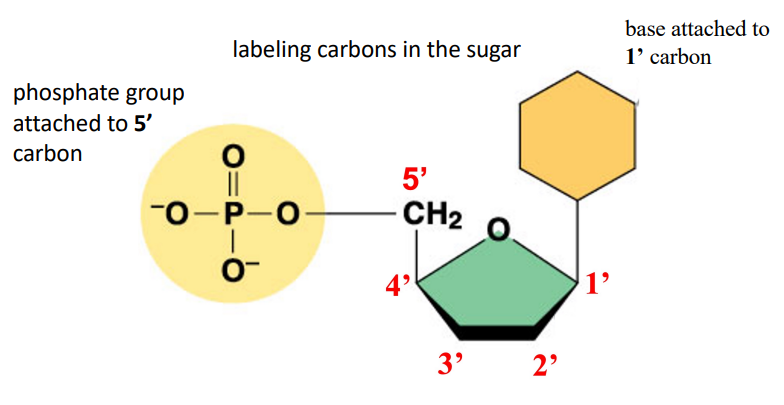
The sugar is attached to the first carbon, and the 5th carbon is attached to the phosphate group. Oxygen separates the first and fourth carbon
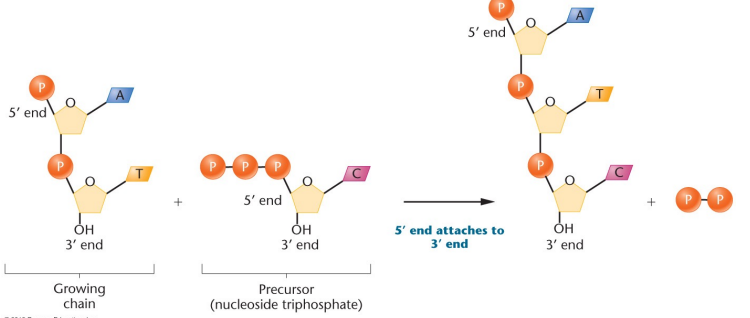
New nucleotides are always added to the 3’ end
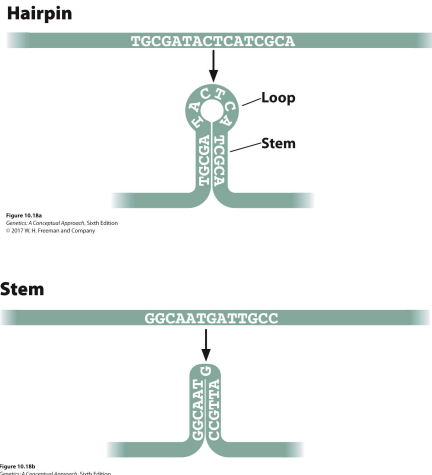
Secondary structures of RNA that allow it to fold into different shapes, like if RNA matches it own base pair later on it may fold