Chapter 16: Amino Acids, Proteins, and Enzymes
16.1: Proteins and Amino Acids
Protein
- Structural Protein: It provides structural components.
- Contractile Protein: It makes muscle moves.
- Transport Proteins: These carry essential substances throughout the body
- Storage Proteins: These store nutrients.
- Hormone Proteins: These regulate body metabolism and the nervous system.
- Enzyme Proteins: These catalyze biochemical reactions in the cells.
- Protection Proteins: These recognize and destroy protein substances.
Amino Acids
These are the molecular building blocks of proteins.
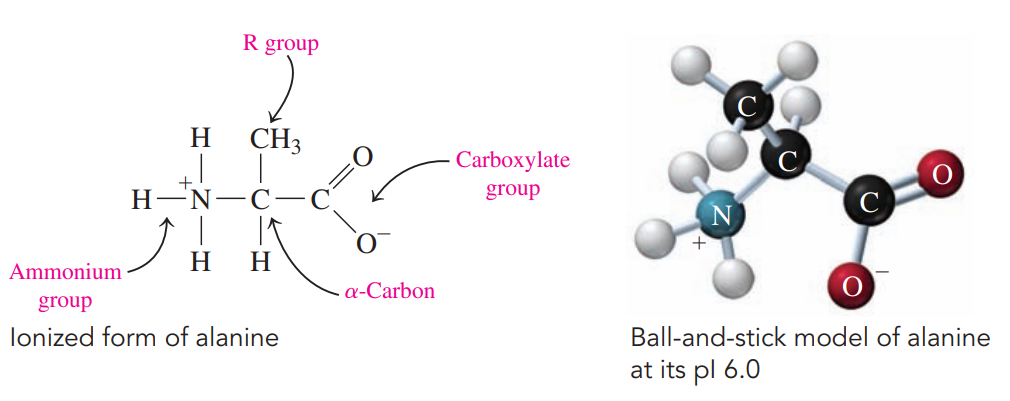
Every amino acid has a central carbon atom called the ɑ-carbon bonded to two functional groups:
An ammonium group
A carboxylate group
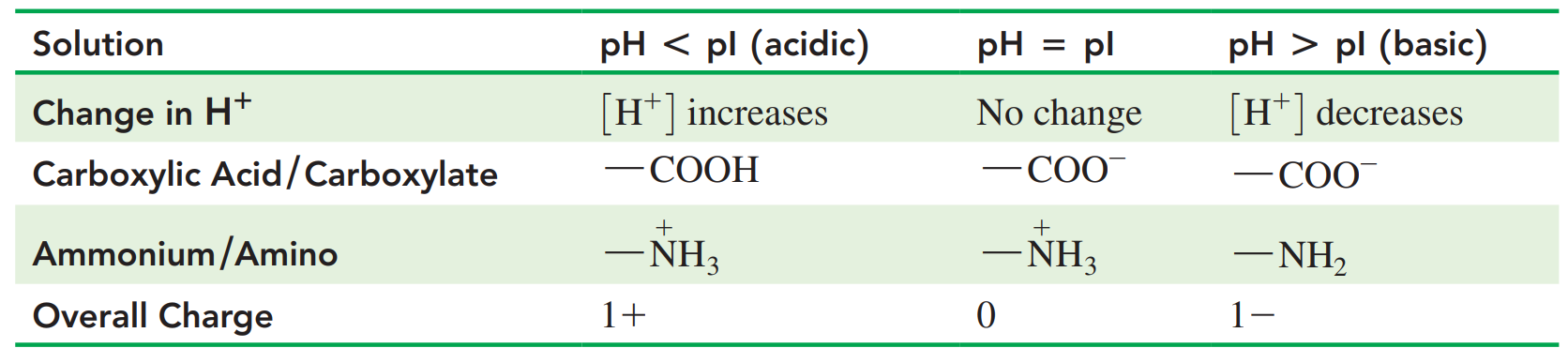
Isoelectric Point: The pH at which an amino acid exists in an ionized form with an overall net charge of zero.
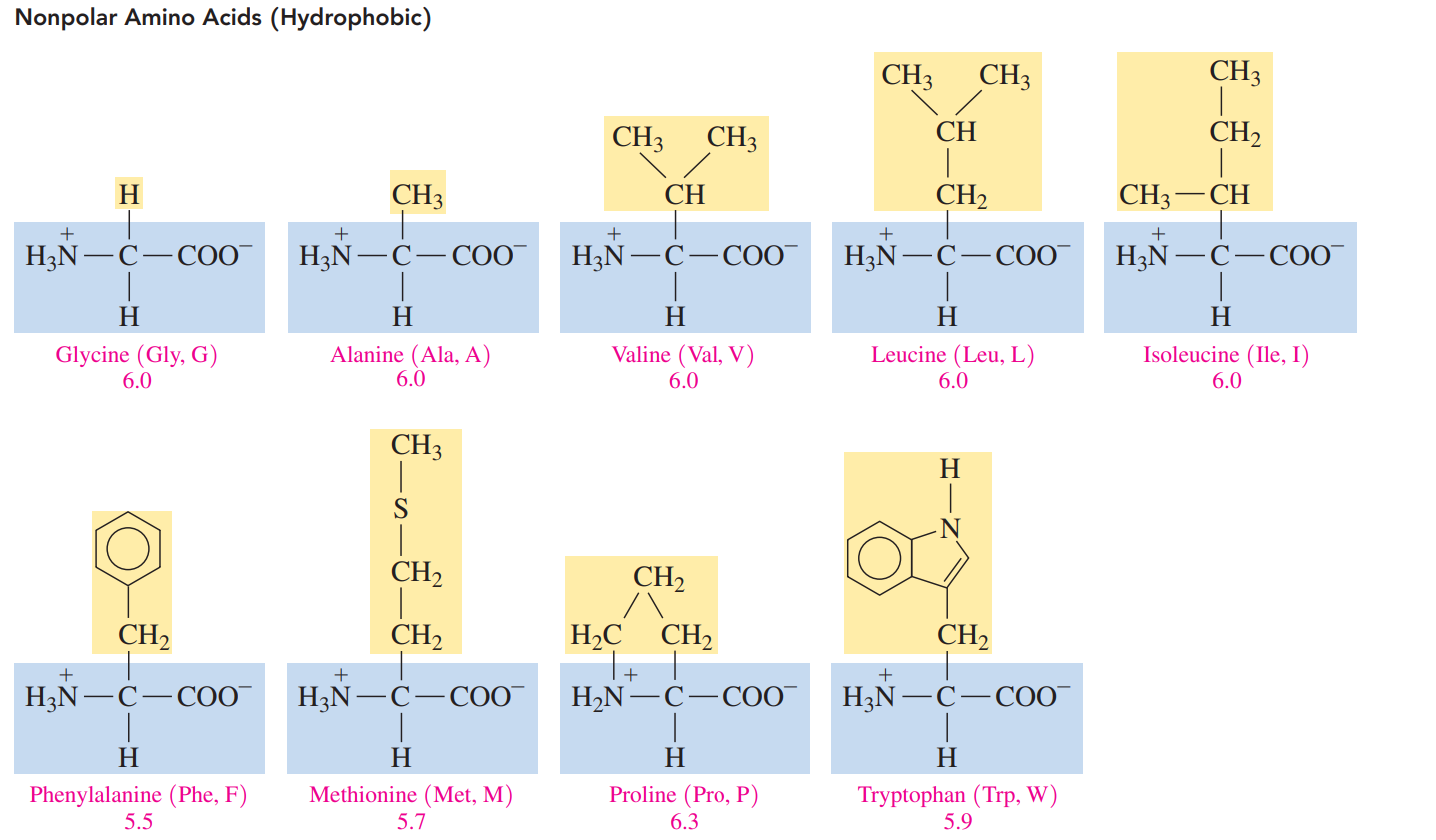
Nonpolar amino acids: These acids have hydrogen, alkyl, or aromatic R groups, which make them hydrophobic.
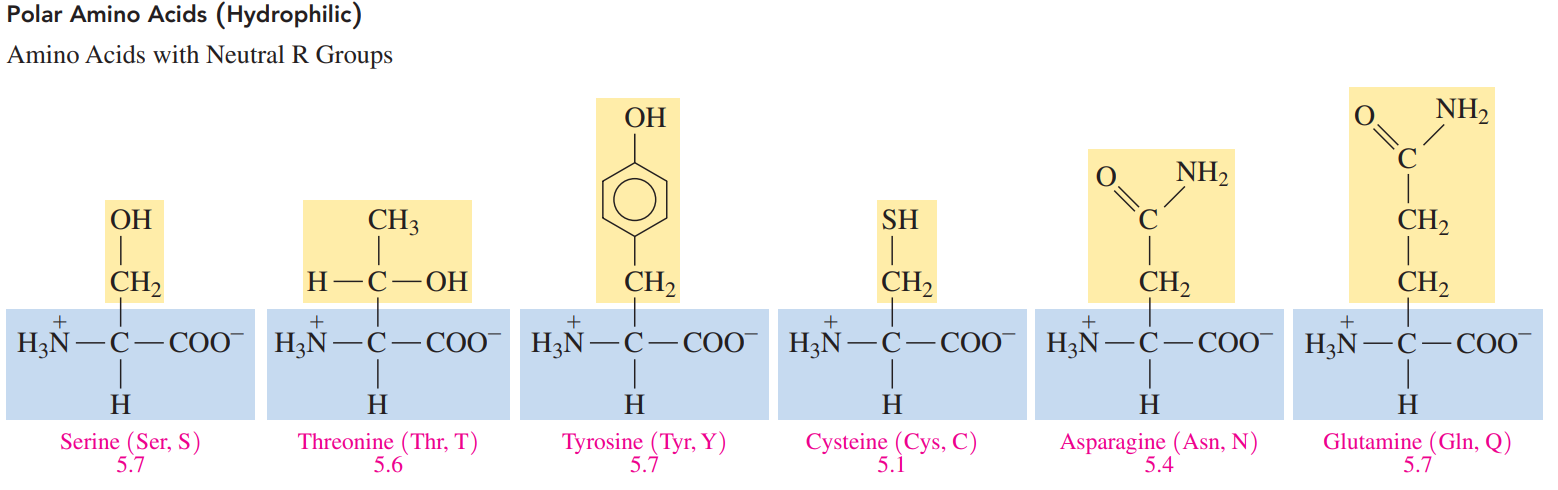
Polar amino acids: These acids have R groups that interact with water, which makes them hydrophilic.
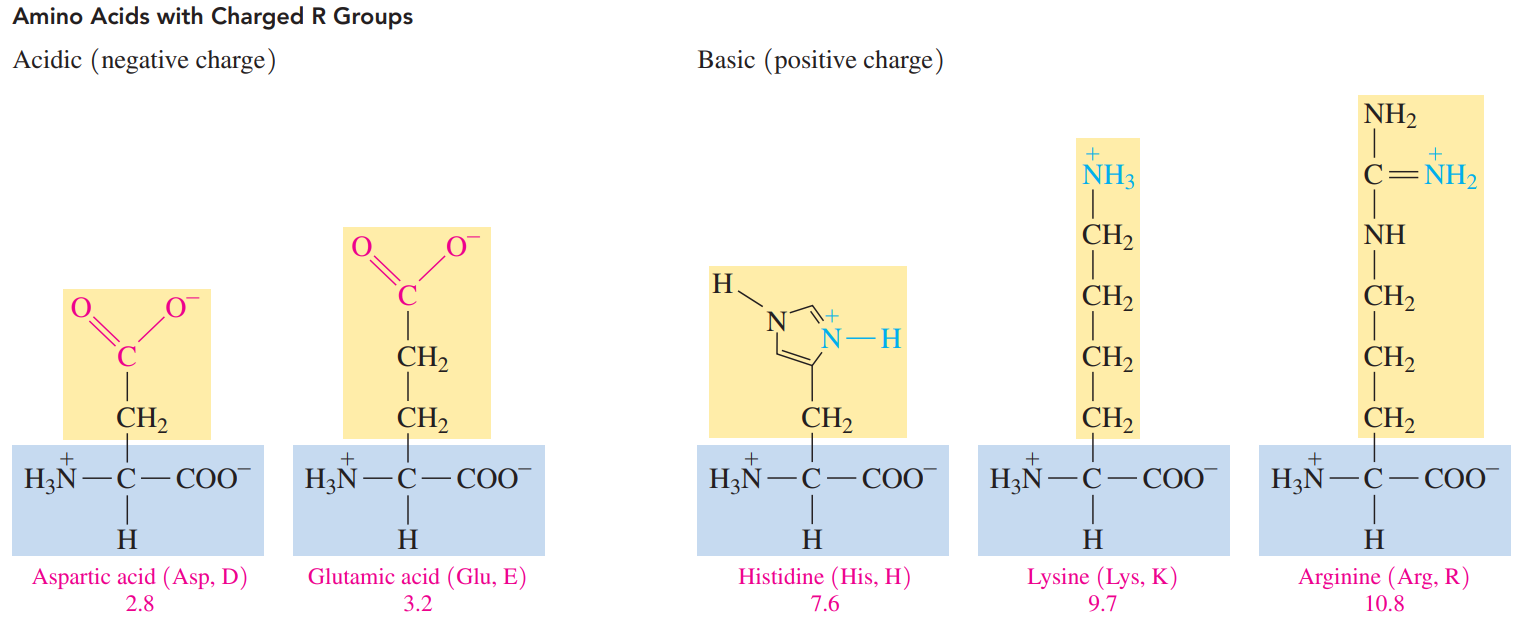
The R group of a polar acidic amino acid contains a carboxylate group
The R group of a polar basic amino acid contains an amino group, which ionizes to give an ammonium ion.
16.2: Amino Acids as Acids and Bases
- An amino acid with positive and negative charges and thus an overall neutral charge forms only at a certain pH called the isoelectric point (pI).
- The pI values for nonpolar and polar neutral amino acids are from pH 5.1 to 6.3. L.
- The pI values of the acidic amino acids are around pH 3, and the carboxylic acid group in the R groups is not ionized.
- The pI values of basic amino acids are typically higher than physiological pH values, ranging from pH 7.6 to 10.8.
Ionized Forms of Nonpolar and Polar Neutral Amino Acids
16.3: Proteins: Primary Structures
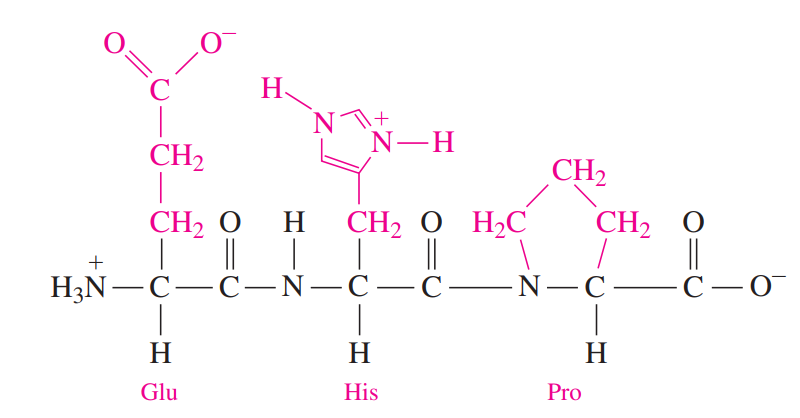
Peptide Bond: An amide bond forms when the —COO- group of one amino acid reacts with the —NH3+ group of the next amino acid.
The linking of two or more amino acids by peptide bonds forms a peptide.
Two amino acids form a dipeptide, three amino acids form a tripeptide, and four amino acids form a tetrapeptide. A chain of five amino acids is a pentapeptide, and longer chains of amino acids form polypeptides.
With the exception of the C-terminal amino acid, the names of all the other amino acids in a peptide end with –yl.
Protein: A polypeptide of 50 or more amino acids that have biological activity.
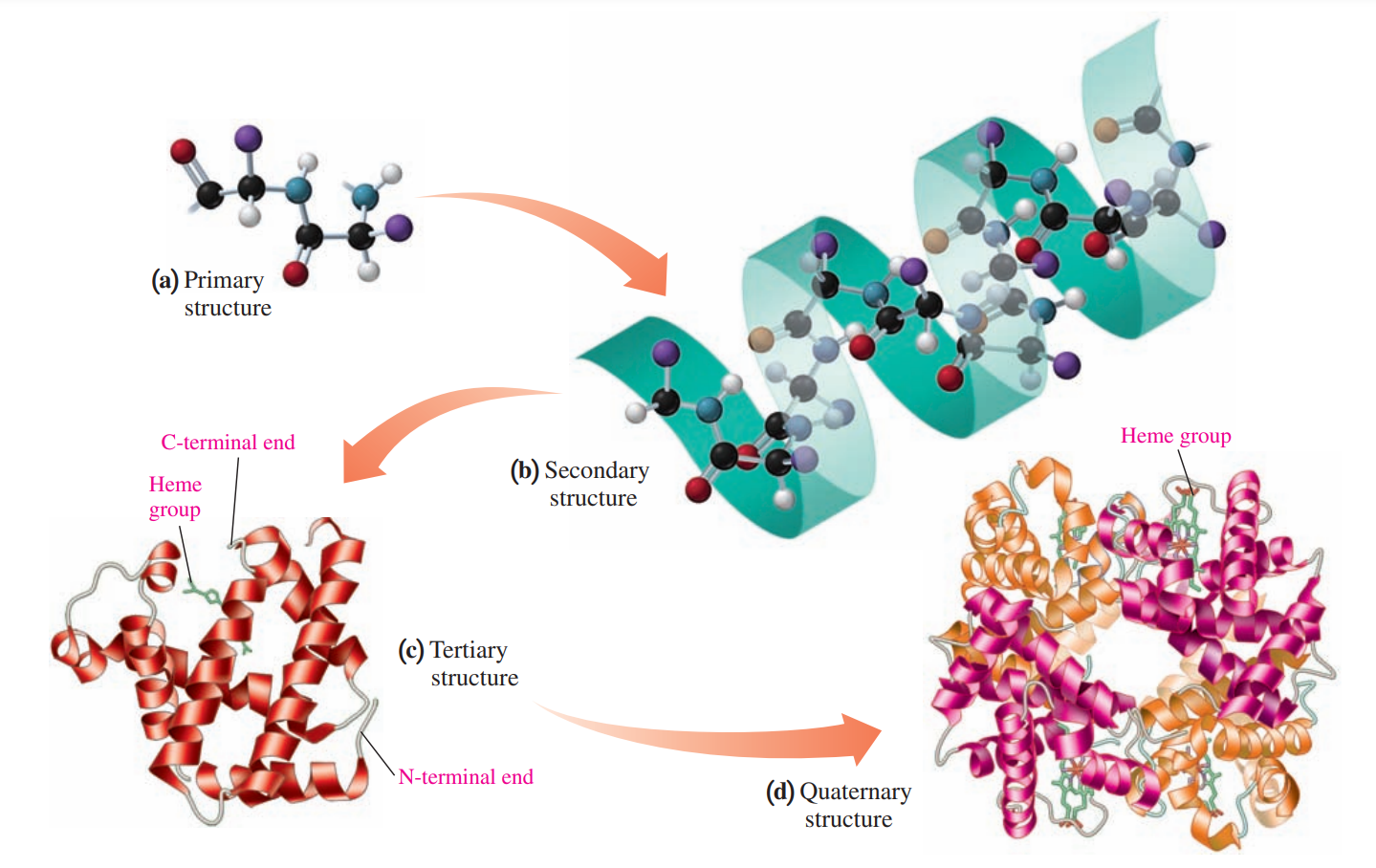
Primary Structure of Protein
- The particular sequence of amino acids is held together by peptide bonds.
- The first protein to have its primary structure determined was insulin, which was accomplished by Frederick Sanger in 1953.
16.4: Proteins: Secondary, Tertiary, and Quaternary Structures
Secondary Structure of Protein
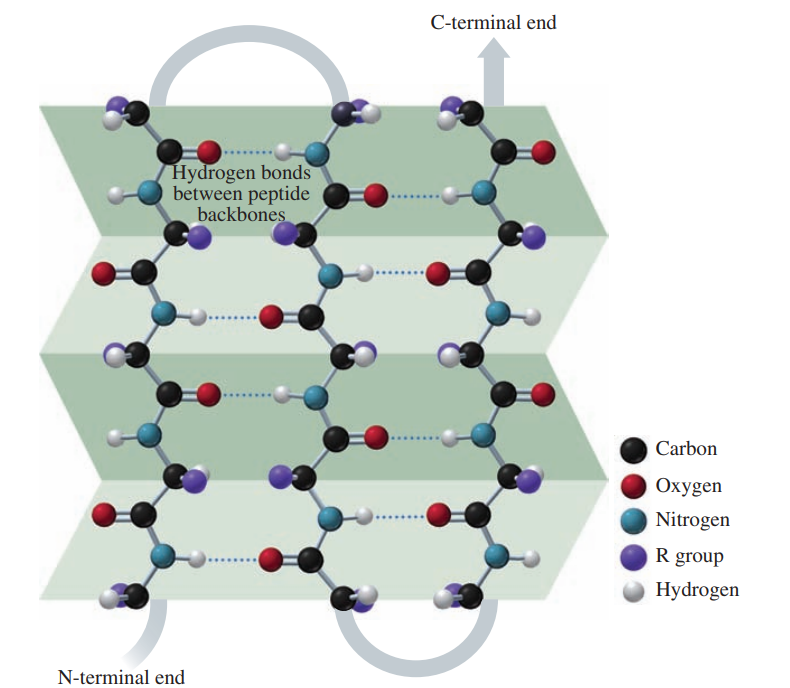
The secondary structure of a protein describes the type of structure that forms when amino acids form hydrogen bonds within a polypeptide or between polypeptide chains.
In an alpha helix, hydrogen bonds form between the oxygen of the C=O groups and the hydrogen of N—H groups of the amide bonds in the next turn of the ɑ helix.
In a beta-pleated sheet, hydrogen bonds form between the oxygen atoms in the carbonyl groups in one section of the polypeptide chain and the hydrogen atoms in the N—H groups of the amide bonds in a nearby section of the polypeptide chain.
Collagen: It makes up as much as one-third of all proteins in vertebrates. It is found in connective tissue, blood vessels, skin, tendons, ligaments, the cornea of the eye, and cartilage.
The strong structure of collagen is a result of three polypeptides woven together like a braid to form a triple helix.
Tertiary Structure of Protein
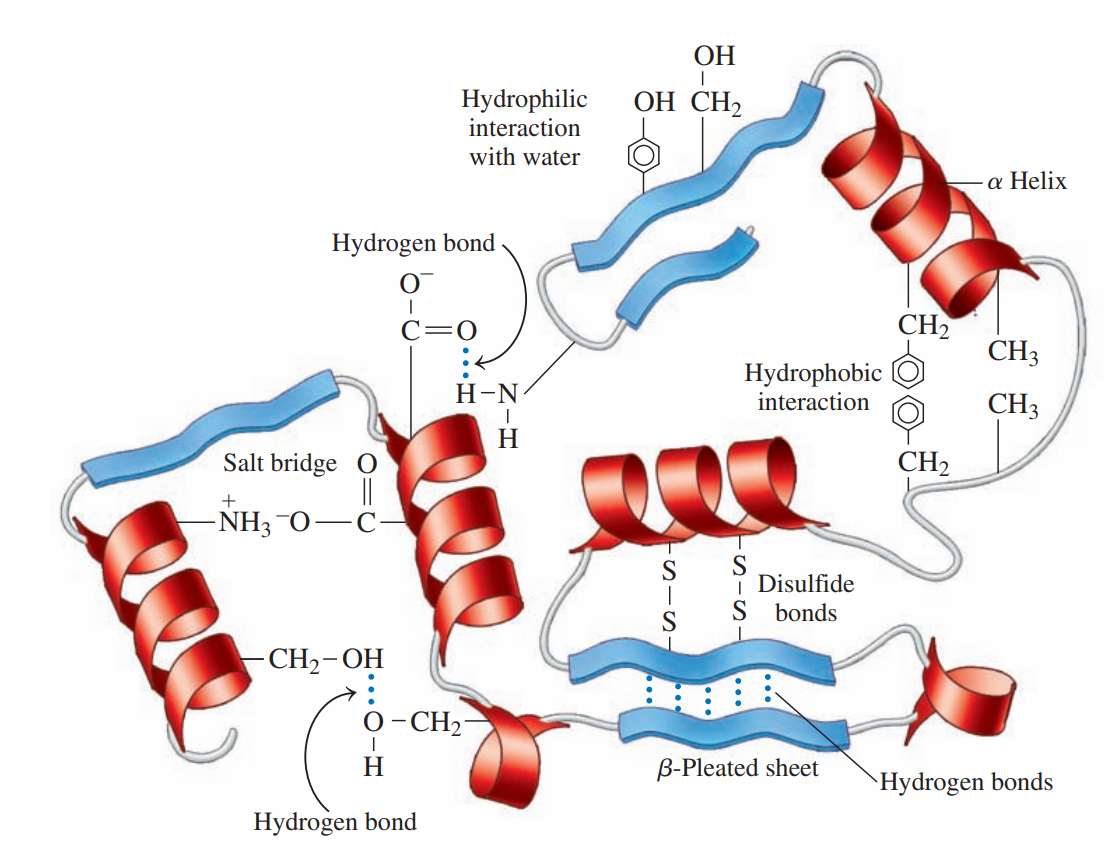
The folding of the secondary structure of a protein into a compact structure that is stabilized by the interactions of R groups.
Hydrophobic interactions: These are interactions between two nonpolar amino acids that have nonpolar R groups.
Hydrophilic interactions: These are attractions between the external aqueous environment and the R groups of polar amino acids that are pulled to the outer surface of globular proteins where they form hydrogen bonds with water.
Salt bridges: These are ionic bonds between ionized R groups of basic and acidic amino acids.
Hydrogen bonds: These form between the H of a polar R group and the O or N of another amino acid.
Disulfide bonds: These are covalent bonds that form between the —SH groups of cysteines in a polypeptide chain.
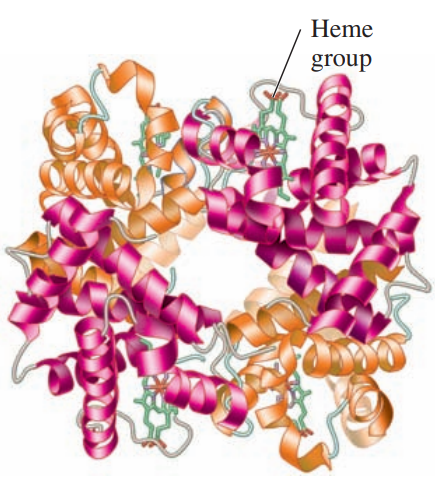
Quaternary Structure: Hemoglobin
A protein structure in which two or more protein subunits form an active protein.
Hemoglobin: A globular protein that transports oxygen in the blood, consists of four polypeptide chains:
two 𝛼-chains with 141 amino acids, and
two 𝜷-chains with 146 amino acids.
In the quaternary structure, the subunits are held together by the same interactions that stabilize tertiary structures.
Each subunit of the hemoglobin contains a heme group that binds oxygen.
Myoglobin: A single polypeptide chain with a molar mass of 17 000, has about one-fourth the molar mass of hemoglobin.
Globular Proteins: Group of proteins that have compact, spherical shapes because sections of the polypeptide chain fold over on top of each other due to the various interactions between R groups.
Fibroid Proteins: These are proteins that consist of long, thin, fiber-like shapes.
- They are typically involved in the structure of cells and tissues.
- 𝛼-keratins: These are the proteins that makeup hair, wool, skin, and nails. In hair, three helices coil together like a braid to form a fibril.
- 𝜷-keratins: These are the type of proteins found in the feathers of birds and scales of reptiles.
Denaturation of Proteins
- It occurs when there is a change that disrupts the interactions that stabilize the secondary, tertiary, or quaternary structure.
- It also occurs by adding certain organic compounds or heavy metal ions or by mechanical agitation.
16.5: Enzymes
Enzymes: These are biological catalysts that are needed for most chemical reactions that take place in the body.
Catalyst: It increases the reaction rate by changing the way a reaction takes place but is itself not changed at the end of the reaction.
An uncatalyzed reaction in a cell may take place eventually, but not at a rate fast enough for survival.
The actual names of enzymes are derived by replacing the end of the name of the reaction or reacting compound with the suffix –ase.
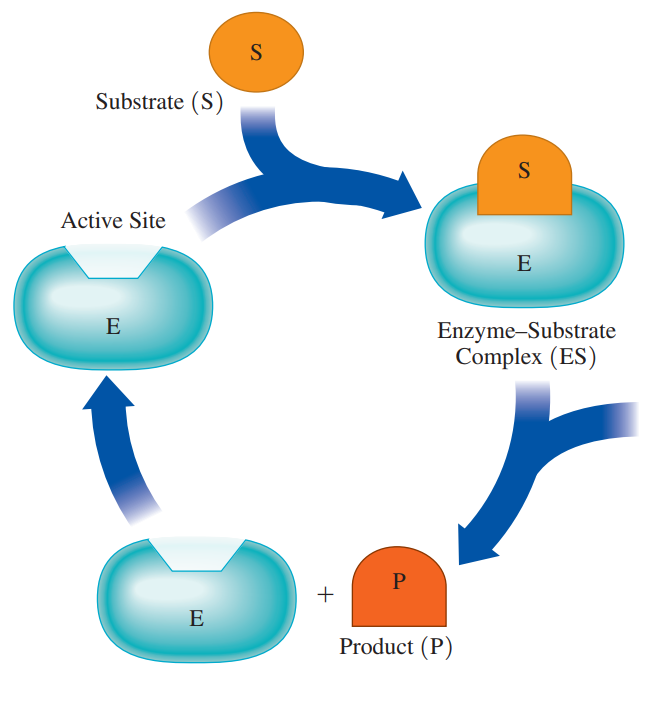
Substrates: The molecule that reacts in the active site in an enzyme-catalyzed reaction.
Active site: A pocket in a part of the tertiary enzyme structure that binds substrate and catalyzes a reaction.
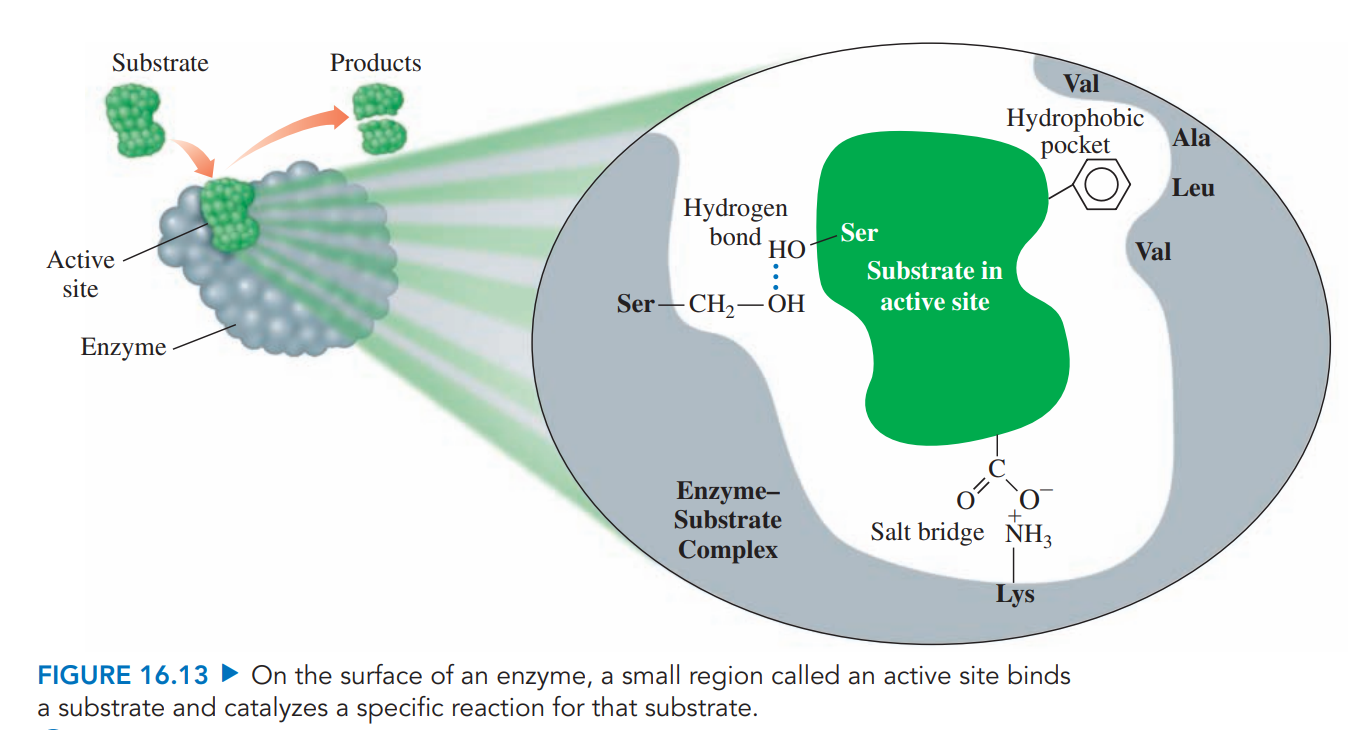
Enzyme–Substrate Complex: It is formed when there’s a combination of an enzyme and a substrate within the active site that provides an alternative pathway with lower activation energy.
Lock-And-Key Model
- It described the active site as having a rigid, nonflexible shape.
- According to this theory, the shape of the active site was analogous to a lock, and its substrate was the key that specifically fit that lock.
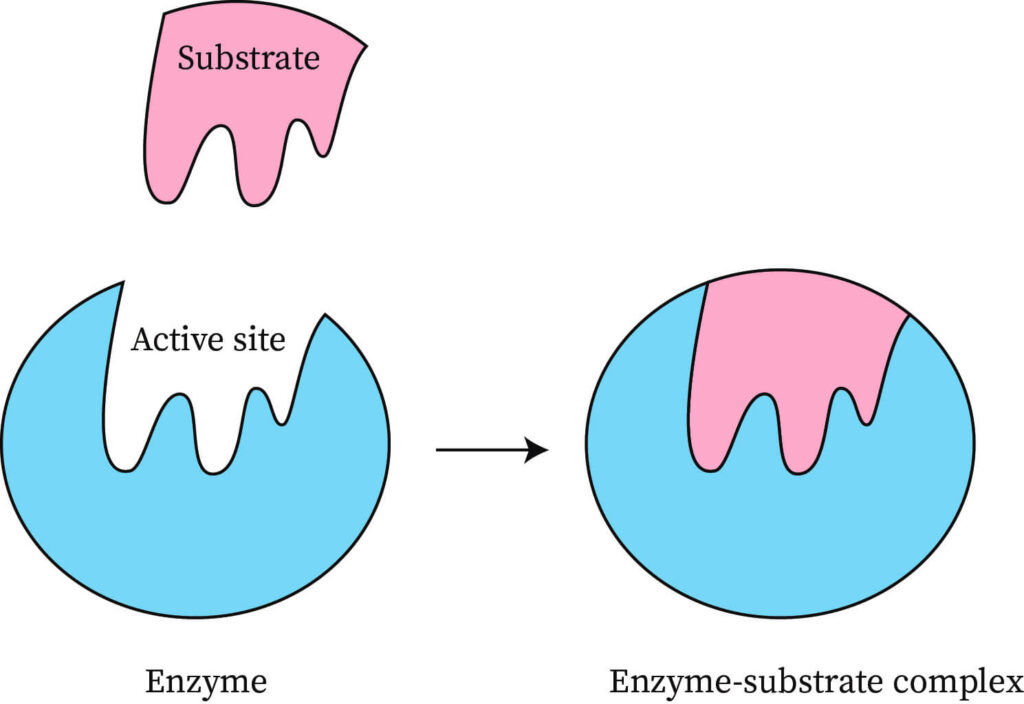
Induced-Fit Model
- In the dynamic model of enzyme action, the flexibility of the active site allows it to adapt to the shape of the substrate.
- At the same time, the shape of the substrate may be modified to better fit the geometry of the active site.
Classes of Enzymes
- Oxidoreductases: For oxidation–reduction reactions.
- Oxidases oxidize a substance.
- Reductases reduce a substance.
- Dehydrogenases remove 2H atoms to form a double bond.
- Transferases: Transfer a group between two compounds
- Transaminases transfer amino groups.
- Kinases transfer phosphate groups.
- Hydrolases: For hydrolysis reactions.
- Proteases hydrolyze peptide bonds in proteins.
- Lipases hydrolyze ester bonds in lipids.
- Carbohydrases hydrolyze glycosidic bonds in carbohydrates.
- Phosphatases hydrolyze phosphodiester bonds.
- Nucleases hydrolyze nucleic acids.
- Lyases: Add or remove groups involving a double bond without hydrolysis.
- Carboxylases add CO2.
- Deaminases remove NH3.
- Isomerases: Rearrange atoms in a molecule to form an isomer.
- Isomerases convert cis to trans isomers or trans to cis isomers.
- Epimerases convert D to L stereoisomers or L to D.
- Ligases: Form bonds between molecules using ATP energy.
- Synthetases combine two molecules.
16.6: Factors Affecting Enzyme Activity
- The activity of an enzyme describes how fast an enzyme catalyzes the reaction that converts a substrate to a product.
- Temperature
- Enzymes are very sensitive to temperature.
- At low temperatures, most enzymes show little activity because there is not a sufficient amount of energy for the catalyzed reaction to take place.
- At higher temperatures, enzyme activity increases as reacting molecules move faster to cause more collisions with enzymes.
- Enzymes are most active at optimum temperature, which is 37 °C, or body temperature, for most enzymes.
- pH
- Enzymes are most active at their optimum pH, the pH that maintains the proper tertiary structure of the protein.
- If a pH value is above or below the optimum pH, the R group interactions are disrupted, which destroys the tertiary structure and the active site. As a result, the enzyme no longer binds the substrate, and no catalysis occurs.
- Enzyme Inhibition
- Inhibitors: Kinds of molecules that cause enzymes to lose catalytic activity.
- An enzyme with a reversible inhibitor can regain enzymatic activity, but an enzyme attached to an irreversible inhibitor loses enzymatic activity permanently.
- Competitive Inhibitor: It has a chemical structure and polarity that is similar to that of the substrate.
- Noncompetitive Inhibitor: It does not resemble the substrate and does not compete for the active site. It binds to a site on the enzyme that is not the active site.
- Irreversible Inhibition
- In irreversible inhibition, a molecule causes an enzyme to lose all enzymatic activity. Most irreversible inhibitors are toxic substances that destroy enzymes.
- It forms a covalent bond with an amino acid side group within the active site, which prevents the substrate from binding to the active site or prevents catalytic activity.