M1L4 Mechanisms of transcription and impact of its dysregulation in cancer
There are ~21,000 protein coding genes in the genome (3-5%)
3 RNA polymerases (I, II, III)
Pol II transcribes protein coding genes (10%)
Pol I transcribes rRNA + Pol III transcribes tRNA (90%)
Regulatory DNA elements
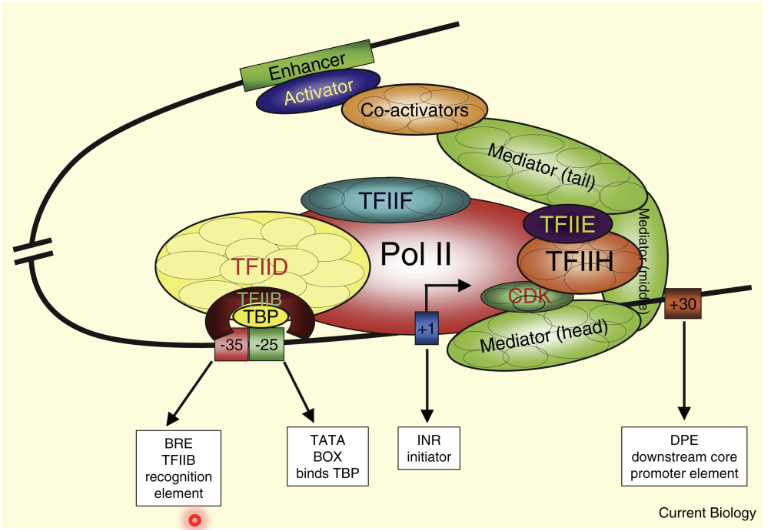
TSS (transcription start site) at +1, where Pol II binds
Transcription factors (TFIID, TFIIB, TFIIE, THFIIH)
Upstream of TSS - TATA box at -25 or -35 which binds TBP and contains TFIIB recognition element
Downstream DPE core promoter element
Enhancers can be located close or far away from the promoter on the same chromosome or even on a different chromosome
Brought to close proximity to promoter by looping the DNA (same chromosome) or bringing close in trans (different chromosome)
Activators, co-activators and mediators assist in this
Insulated sequences (tight chromatin) - prevents certain enhancers from acting on the wrong DNA sequences, eg proto oncogenes
Enhancer DNA sequences are also transcribed, so they may not just act as DNA elements but can also act as scaffold and have regulatory functions as transcribed elements
Transcription is the combinatorial action of DNA cis elements located close to the TSS and trans elements like enhancers, TFs etc.
Transcription is highly regulated
PIC (pre-initiation complex) step wise assembly
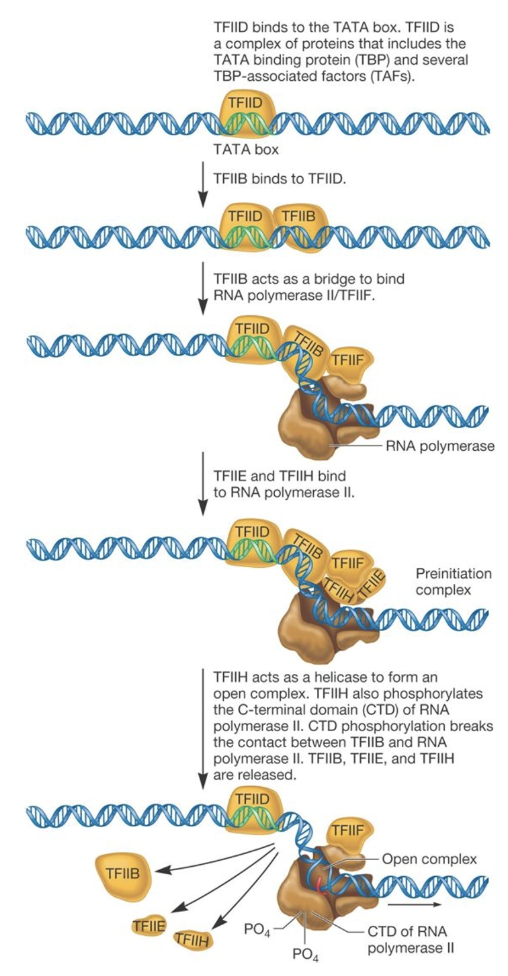
TFIID (complex of proteins containing TBP and TBP-associated factors (TAFs)) binds to TATA box
TFIIB recruited and acts as a bridge with RNA pol II
TFIIE and TFIIH bind to PNA pol II, forming the PIC
TFIIH helicase activity forms an open complex and it also phosphorylates the RNA pol II CTD, breaking the contact between TFIIB and RNA pol II and releasing TFIIB, TFIIE and TFIIH
Pol II moves downstream and begins RNA synthesis
This model assumes that DNA is quite static whereas DNA is very dynamic (chromatin conformational changes, moving around, making 3D contacts … etc)
Transcriptional factories
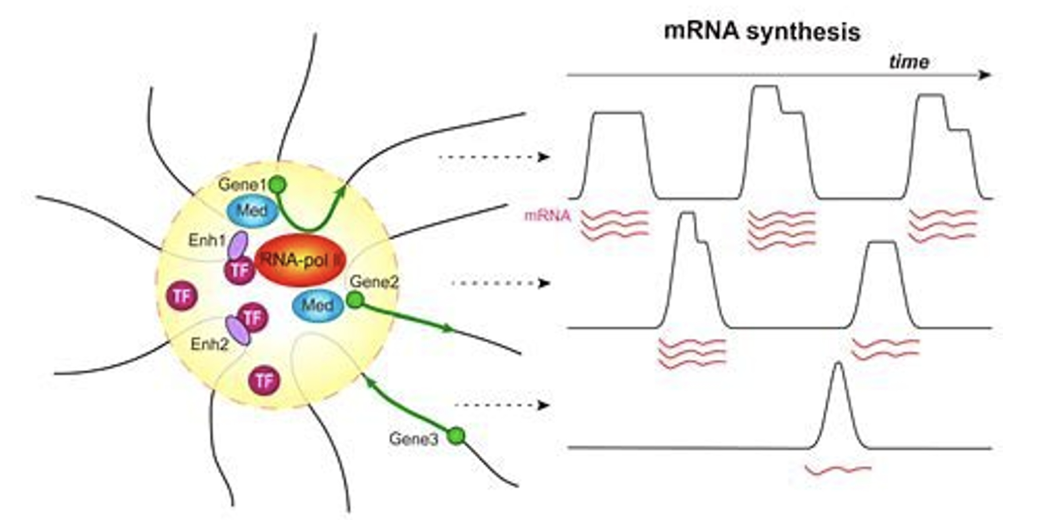
Polymerases concentrated in a space in the nucleus
DNA is brought to a specialised ‘factory’ when needed to be transcribed (eg. Pol II ‘factory’ for transcribing protein-coding genes)
Principles of transcription, types of interactions remain true to previous knowledge
Conserves energy by concentrating machinery in one space
Steps in transcription
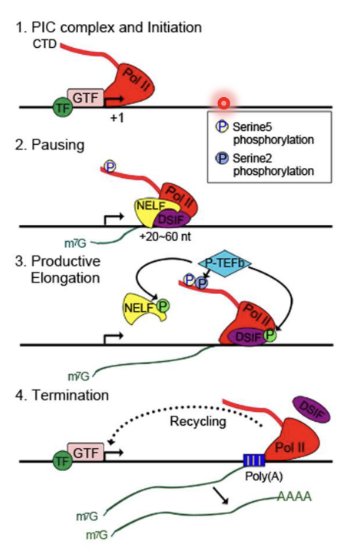
1. PIC complex and initiation - Pol II binds to +1 (TSS) with general TFs
2. Pausing after +20-60nt as NELF and DSIF blocks progression (checkpoint event)

Small proportion progresses after pausing as RNA pol II frequently does not pass the checkpoint, resulting in a dip in the Pol II density
Peak in pol II density in the termination stage is because pol II is slow to disengage
Capping/RNA processing occurs co-transcriptionally
3. Productive elongation - blocking factors removed by phosphorylation by P-TEFb which causes conformational change in NELF (causing it to disengage and leave) and DSIF (which allow it to disengage and travel with RNA pol II)
As RNA is being produced it can also form secondary structures or even invade back into the DNA duplex, this may also be important for regulatory processes (G quadruplex, i-motifs, triplexes, Z-DNA/RNA, DNA cruciform, R-loops, RNA hairpins…)
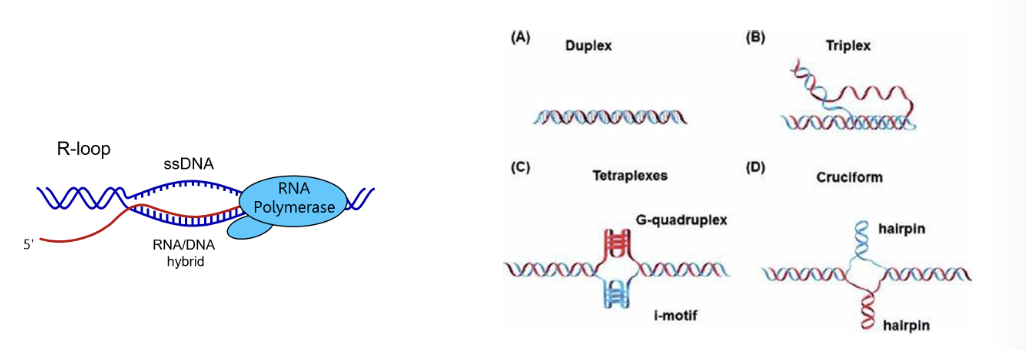
These structures may accumulate in cancer and neurodegeneration
4. Termination - indicated by poly(A) signal on protein coding genes which causes Pol to disengage
Pol II termination
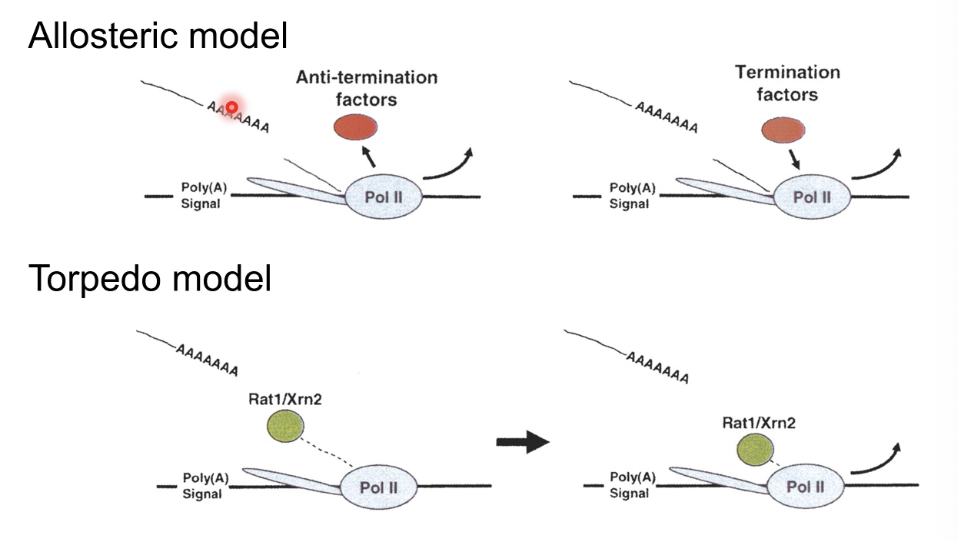
Allosteric model - Poly(A) signal is key, anti-termination factors separate from Pol II and termination factors bind which makes it less processive and causes termination
Torpedo model - After poly(A) site Pol II continues transcription but less efficiently, exonucleases bind to help Pol II to separate from DNA (Rat1/Xrn2 in yeast)
Exonuclease is recruited because there is no p-Ser5 on Pol II CTD which is normally needed for capping, thus the RNA is unprotected which promotes termination
Combined model incorporates both anti-termnination factors and exonucleases
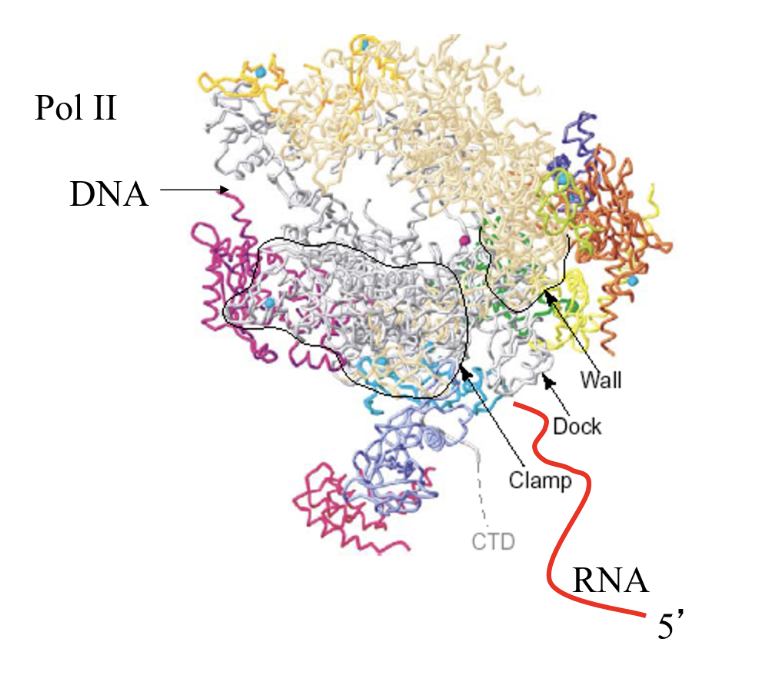
RNA Pol II
12 subunits
CTD - unstructured tail, hanging from big subunit
52 repeats of 7 aa in humans
Changes phosphorylation pattern through different stages of transcription and the signature is repeated many times to create a strong mark (important for pol to move through the stages and coupling with RNA processing)
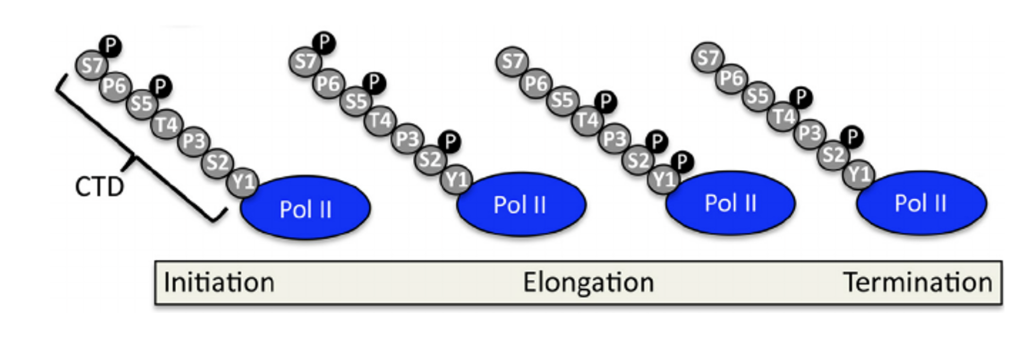
Pol II CTD sits where RNA exists (co-transcriptional RNA processing coupling)
Specialised kinase working with cyclin for phosphorylation
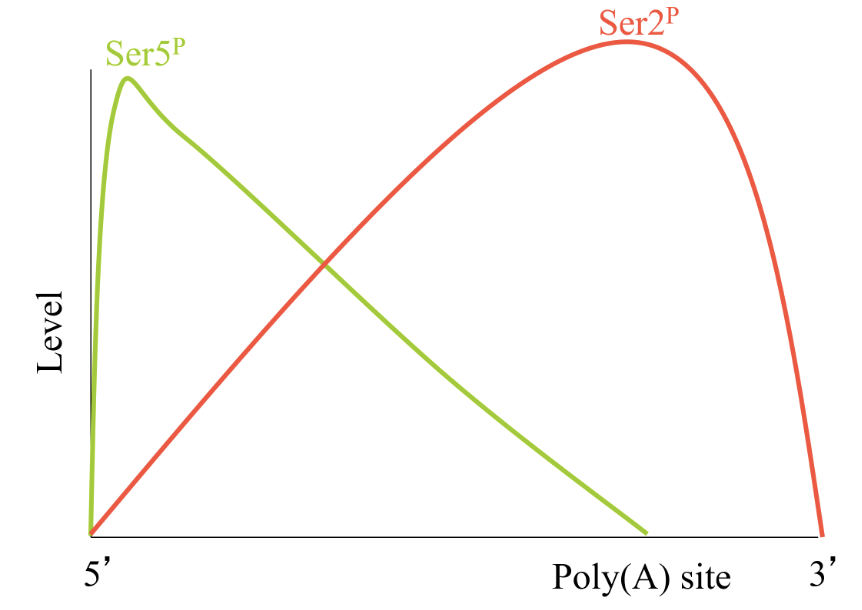
Ser5P is high initially and decreases along the gene
Ser2P is low initially and peaks toward the end
Ser5P recruits capping enzymes (guanylyl transferase)
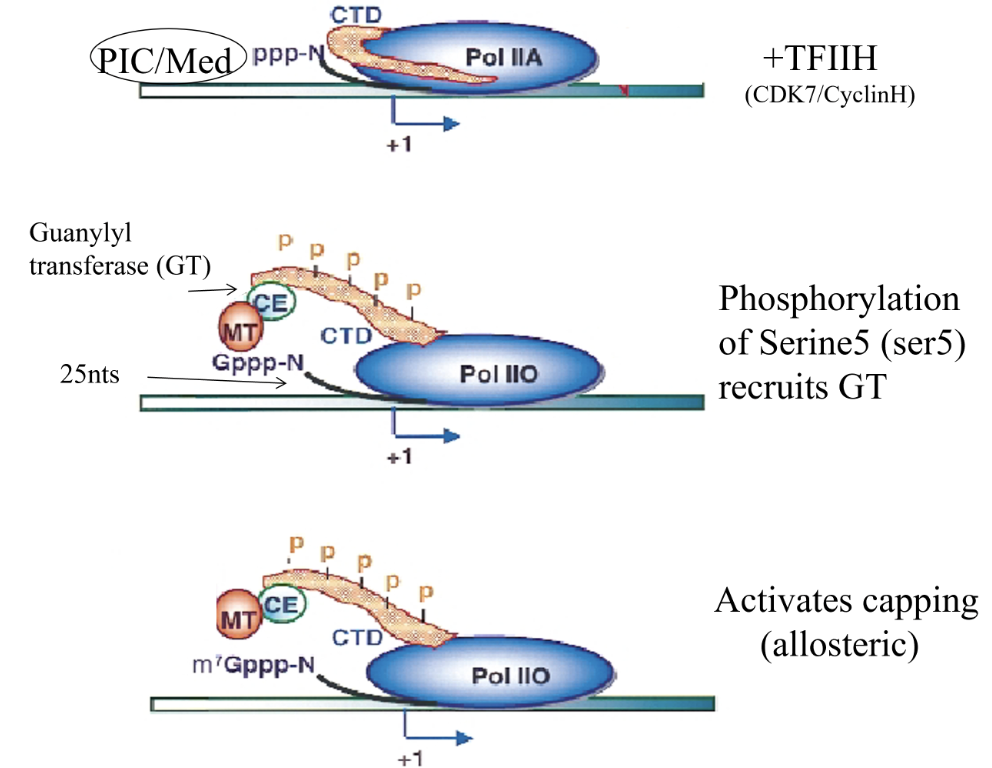
Nascent RNA is capped quickly during early phase of synthesis as a lack of capping triggers degradation by exonucleases (as in termination) and prevents continuation into productive elongation after pausing
Ser5 and 7 are phosphorylated early in transcription by Cdk7/cyclin H (TFIIH)
Can be desphosphorylated by Fcp1
Ser2P recruits splicing factors
Ser2 is phosphorylated by Cdk9/cyclin T1 (P-TEFb) and Cdk12/cyclinK
Dephosphorylated by small CTD phosphatases (SCPs) and Rtr1/RPAP2
After passing the poly(A) site the processed RNA is released but Pol II continues to transcribe for a while, however due to lack of Ser5P, this will cause degradation
Transcription coupled to RNA processing (capping, splicing, polyadenylation)
No progression after pausing without cap
Happens on chroatenised DNA
Transcription and replication happen on same DNA templates
There needs to be balance between replication and transcription for health, imbalance may also be involved in cancer
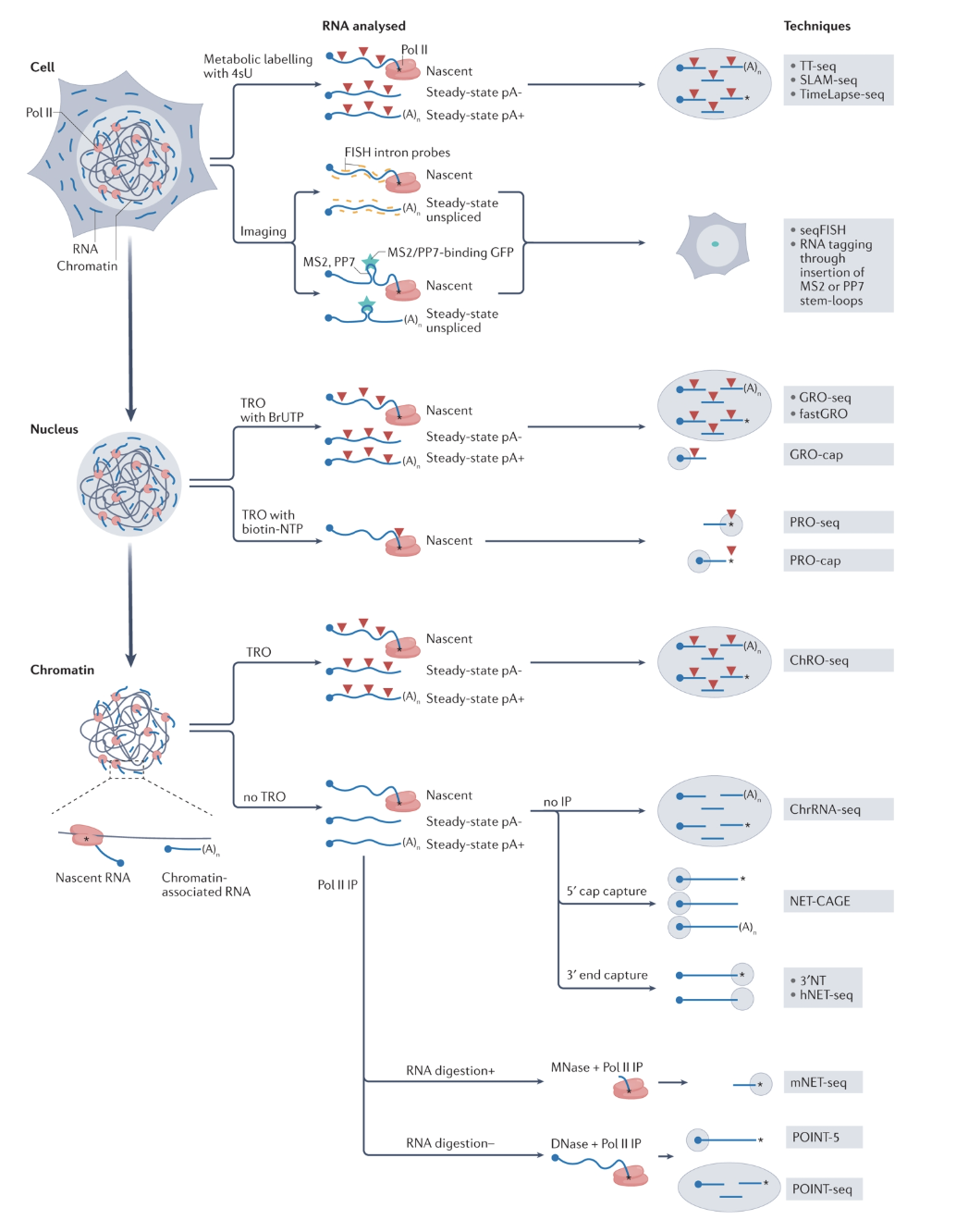
Studying transcription
RT-qPCR only measures mRNA production at the end, rather than nascent RNA production during transcription
Pull down polymerase to sequence nascent RNA as one method to capture nascent RNA production