Chapter 6 Energy and Enzymes
Chapter 6 - Energy and Enzymes
Slides 2-3: Types of Energy in the Cell
Chemical Energy
ATP/GTP
NADH/FADH2
Other high-energy molecules
Ion Gradients
Energy from ion concentration differences across membranes
Solar Energy
Energy harnessed from sunlight
Types of Chemical Energy
ATP/GTP (Most common)
Primary energy carriers in cells
NADH/FADH2
Electron carries in metabolic reactions
Other High Energy Molecules
Various molecules that store energy
Slides 4-5: ATP: Adenosine Triphosphate
Structure of ATP
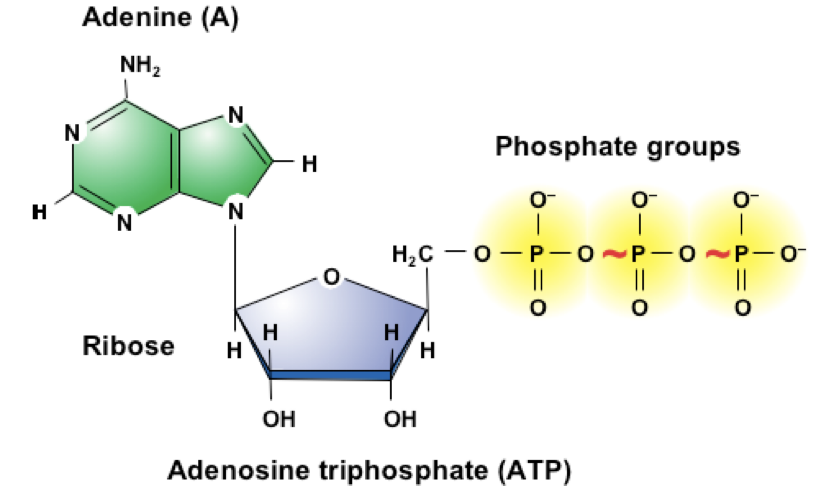
Composed of adenine, ribose, and phosphate groups
ATP has High-energy bonds between the phosphate groups
When the P-P bonds are broken the energy released can be used for enzyme reactions.
Slides 6-7: ATP Hydrolysis
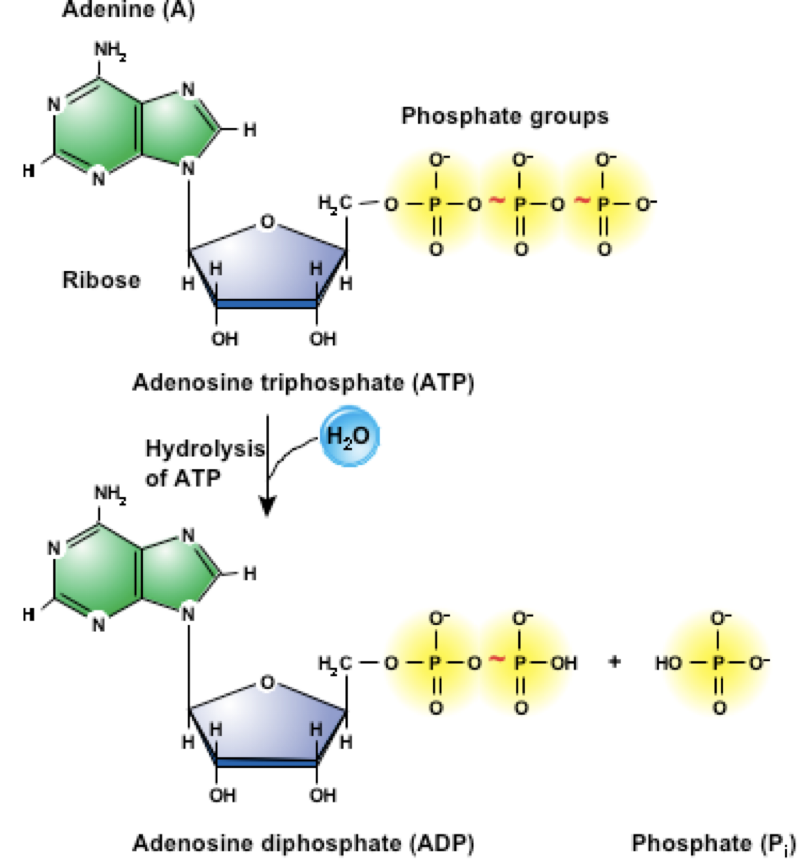
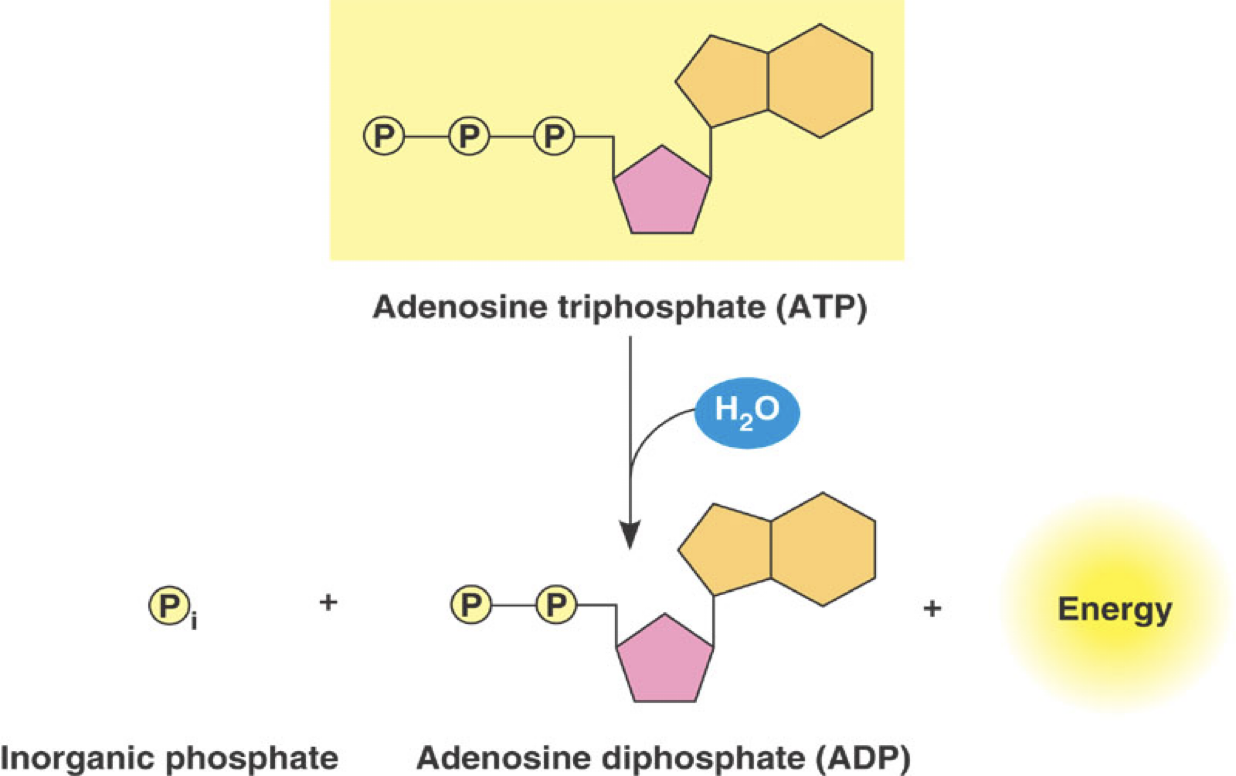
ATP Hydrolysis provides energy needed for many essential process in organisms and cells
Including intracellular signaling, DNA and RNA synthesis and more
Hydrolysis Reaction
ATP + H2O → ADP + Pi + Energy
Energy released can drive cellular processes
Slides 8-16: Electron Carriers: NADH, NADPH, FADH2 —Oxidation-Reduction Reactions —
Redox Reactions (OILRIG)
Oxidation-Reduction reactions
Some molecules alternate between reduced and oxidized forms
Ex. NADH + B -> NAD+ + BH
NADH and NAD+
NADH is the reduced form, high energy that has accepted a pair of electrons
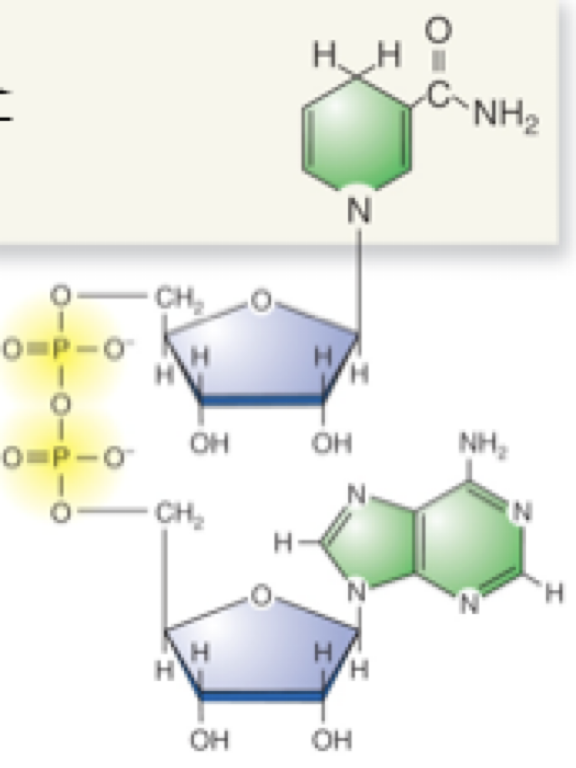
NAD+ is the oxidized form, low energy that has lost a pair of electrons
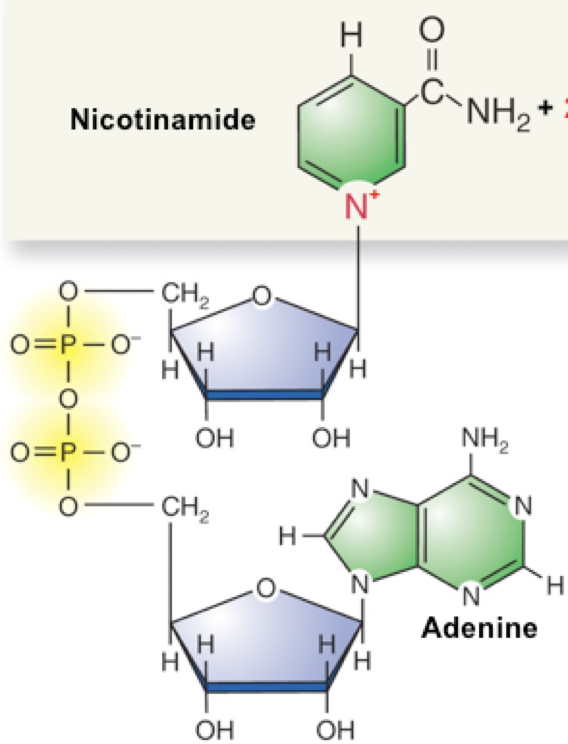
Reduced cmpd loses e-s and becomes oxidized
Oxidized cmpd gains e-s and becomes reduced
When a cmpd gains e-s is sometimes picks up an H as well
NADH (reduced) + B(ox) -> NAD+ (oxidized) +BH(reduced)
The NADH +B donates a pair of e- to B
NADH loses a pair of e- and becomes oxidized — NADH → NAD+
B gains a pair of e- and becomes reduced — B → BH
NAD+ + 2e- + 2H+ -> NADH
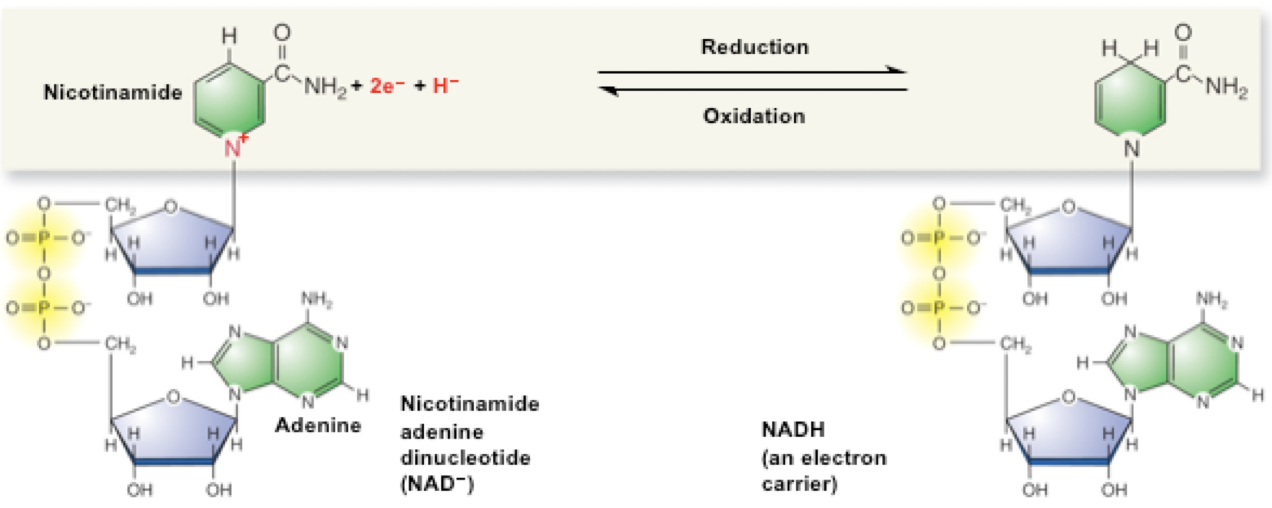
Oxidized form lost a pair of e-s low energy form (left) — Reduced form Gains 2e-s (+ 2H +) high energy form (right)
OILRIG: Oxidation Is Loss, Reduction Is Gain of electrons
Slides 17-19: Ion Gradients as Energy
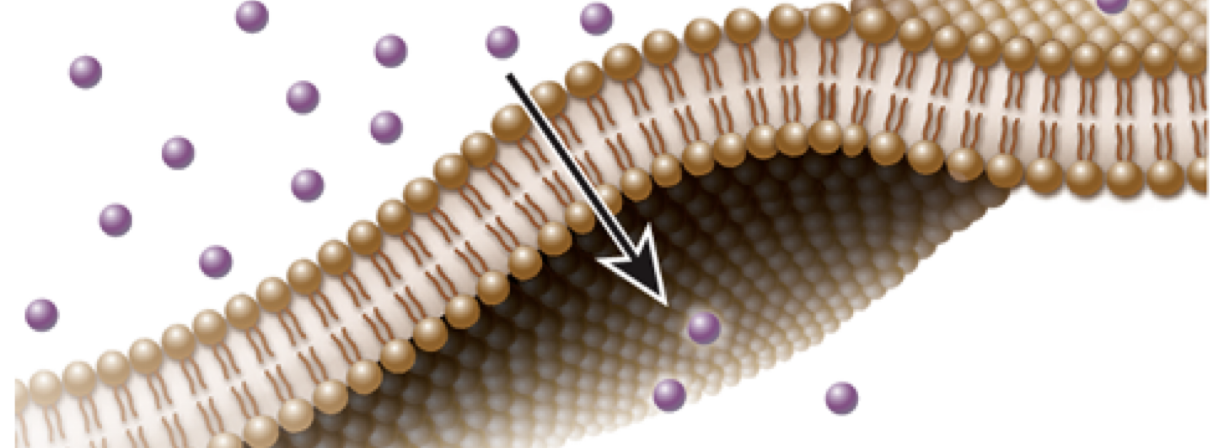
Ion Gradient: When the concentration of an ion is high on one side of the membrane
Ion gradients create potential energy
Energy is released as ions flow down their concentration gradient
As ions move down their concentration gradient, enough energy is released to move some other substance, or make ATP
Slides 20-22: Solar Energy and Photosynthesis
Photosynthesis Overview
Light energy converted into chemical energy (ATP and NADPH)
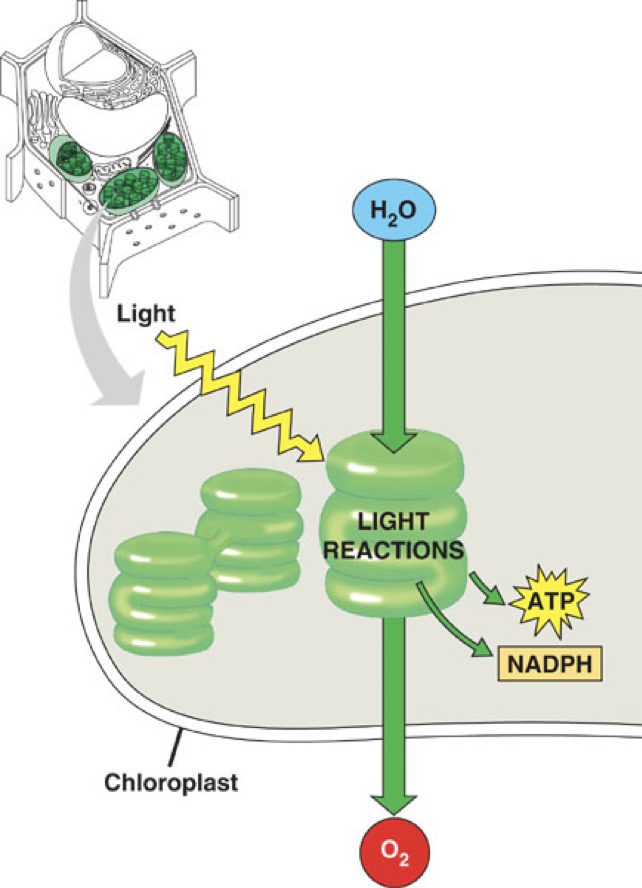
Energy from sunlight can be used for electron transport during photosynthesis
Light Reactions
Involved water splitting and electron transport
Light reactions of photosynthesis — sunlight → Electron transport H+ Gradient → ATP + NADPH chemical energy
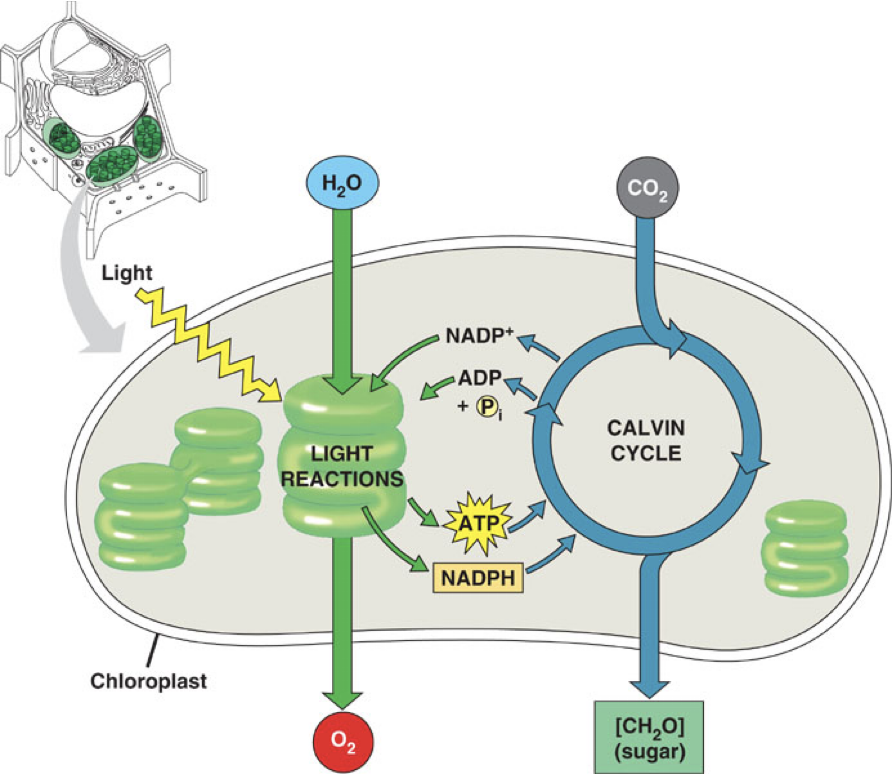
Overall reaction: CO2 + H2O + light energy → sugar + O2
Slides 23-27: Laws of Thermodynamics
1st Law: Energy cannot be created or destroyed, it can only be transformed
2nd Law: Any Energy transformations increases the disorder of a system (entropy)
Entropy is a measure of randomness or disorder
Slides 28-32: Metabolic Pathways and Chemical Reactions
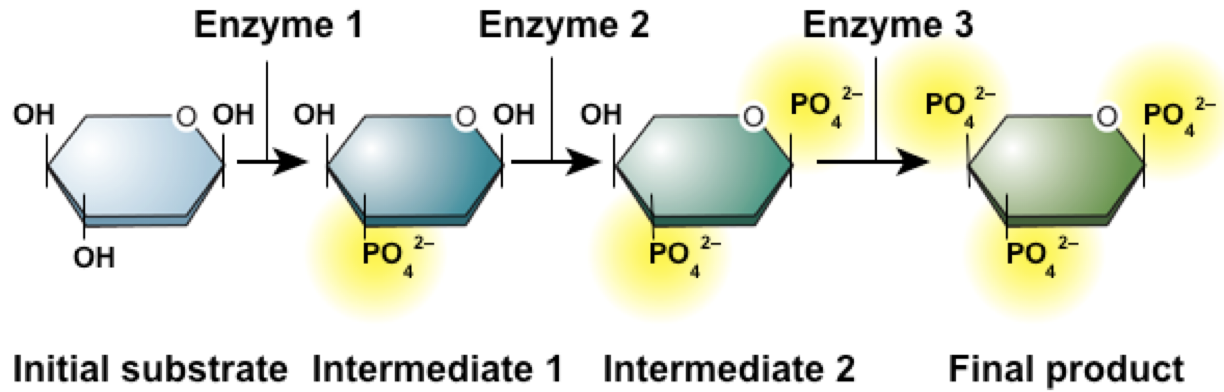
Reactions are part of metabolic pathways
Enzymes catalyze each step
The activity of all 3 enzymes is needed to make the final product
Reactants = substrates is the starting compound in a reaction
Product: Compound made at the end of a reaction
Slides 33-38: Chemical Equilibrium
Chemical equilibrium occurs when the rate of formation of products equals the rate of formation of the reactants
aA + bB <—>cC + dD
Equilibrium constant (Keq) indicates favorability of product formation
The larger the Keq the more product is made relative to reactant at equilibrium
Example: ATP breakdown to ADP + Pi
Equilibrium greatly favors formation of products
Keq = [ADP][Pi] / [ATP]
ATP rapidly breaks down to make ADP + Pi
At equilibrium there will be tiny amounts of ATP and very large amounts of ADP + Pi
Slides 39-44: Free Energy and Reactions
Free energy (G) is the energy that can be used to do work
Reactions tend to proceed in the direction that causes a decrease in the free energy of the system
(ΔG = negative)
Exergonic Reactions: Energy is released can occur spontaneously (ΔG < 0 ) ΔG = negative
Endergonic Reactions: Requires energy input in order to occur reaction is not spontaneous (ΔG > 0) ΔG = Positive
Reactions can be coupled
Coupled reactions over all ΔG is negative; together reactions are spontaneous
ATP → ADP + P
Glucose+ P -: Glucose-6P
Glucose + ATP → Glucose-6-p + ADP
Slides 45-52: Enzymes as Catalysts
Catalysts increase the rate of reaction
Catalysts are not used up in the reaction (reusable!!)
A catalyst does not equal reactant
Catalysts do not alter the equilibrium of a reaction
They decrease the amount of time required to reach equilibrium
Special Features of Enzymes
Enzymes are specific, efficient and regulated
Specific : Enzymes perform one reaction or type of reaction
Efficient : Enzymes increase the rate of reaction dramatically
Regulated : Enzymes can be turned ON and OFF.
Enzymes work in path way that can be interconnected and flow thru a pathway can be regulated
Enzymes are proteins that speed up reactions
Catalysts are not consumed in reactions
Slides 54-57: Mechanism of Enzyme Activity
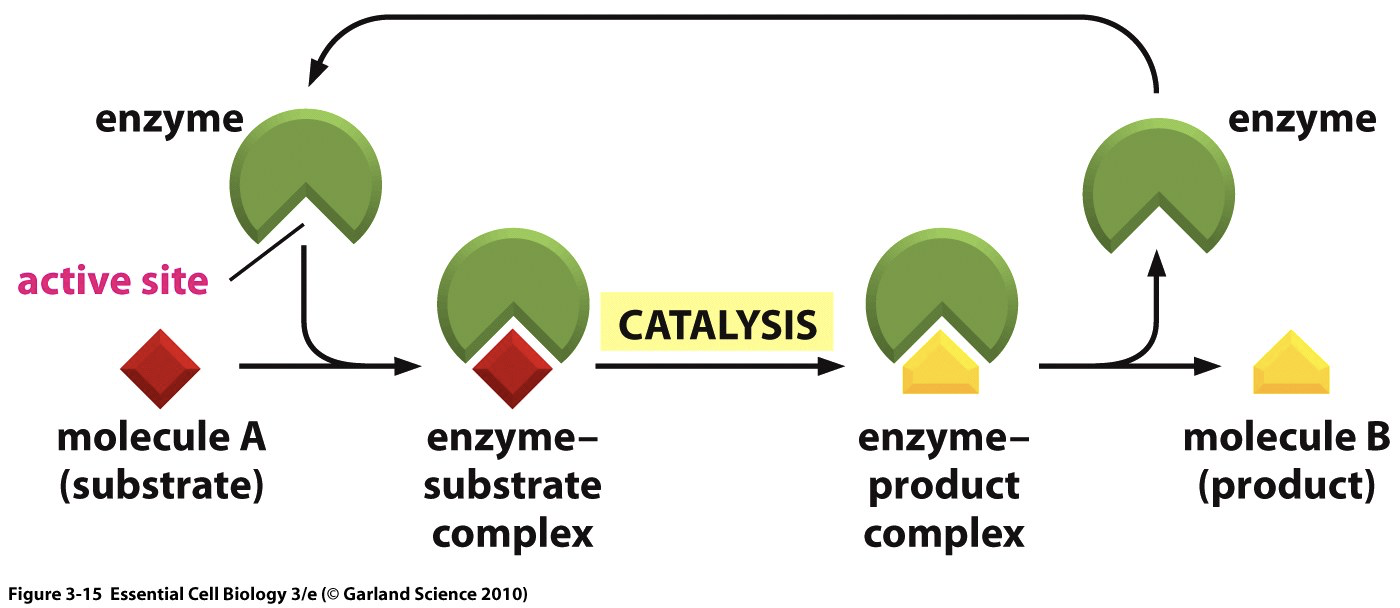
Enzymes interact with substrates to form an enzyme-substrate complex
Transition state is an intermediate with higher free energy (G) compared to that of the reactant
Substrates must go through an activated transition state
E + S -> E.●S -> E.●P -> E + P
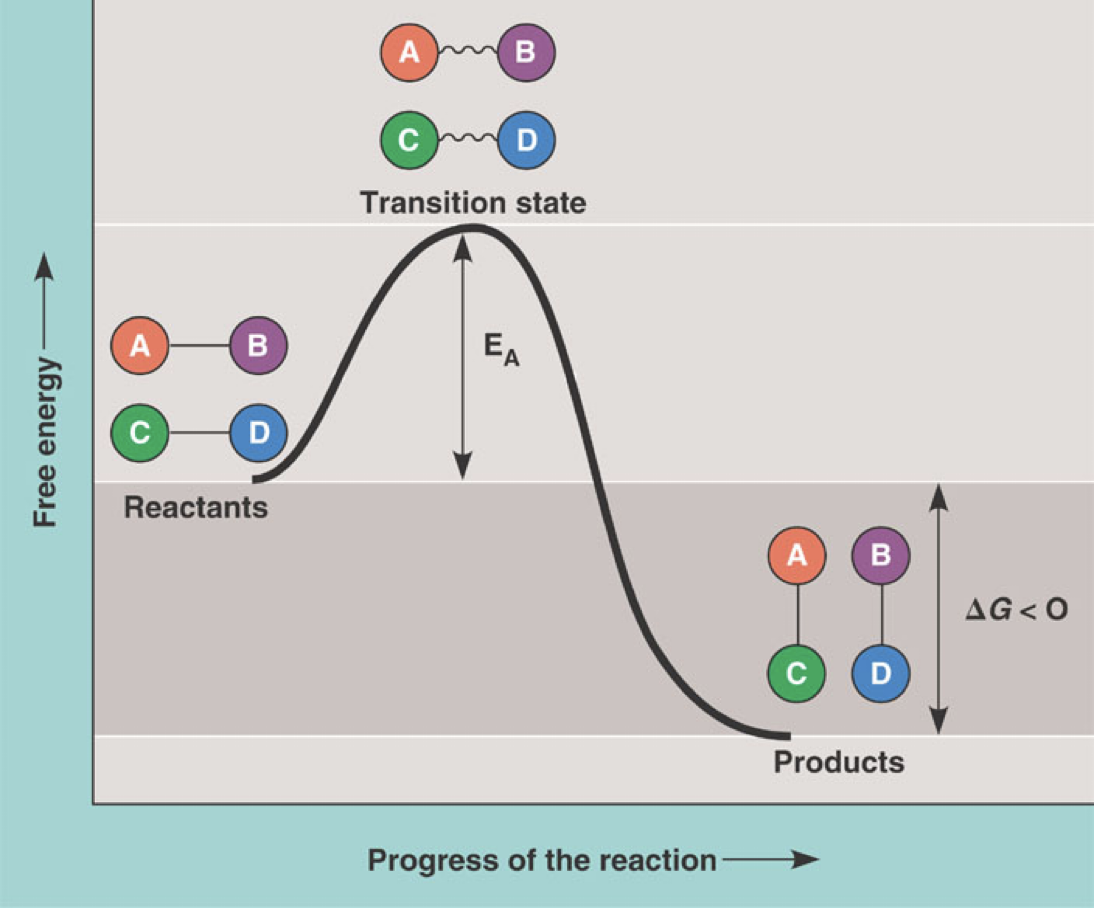
Slides 59-65: Activation Energy
Activation energy (EA) is the energy needed to start a reaction
Enzymes lower the activation energy(EA), speeding up the reactions
Enzymes can interact with the substrate providing an alternate transition state
Alternate state = different intermediate
Enzyme catalyzed reaction goes through a transition state/intermediate whose free energy is lower than the intermediate of the uncatalyzed reaction
To lower the activation energy enzymes can interact with the substrate and provide an alternate transition state
They can also hold the substates in the proper orientation for the reaction to occur
Slides 66-74: Enzyme Specificity and Function
Enzymes specificity is determine by the shape of the active site
Active site: location in enzyme where reaction takes place

Lock and Key Model: Only specific substrates fit
Specificity: only molecules that can fit into the active site of an enzyme are substrates for the reaction
Substrate fit into the active site of an enzyme like a lock into a key
Substrates are oriented so the bond can be cleaved in a certain position
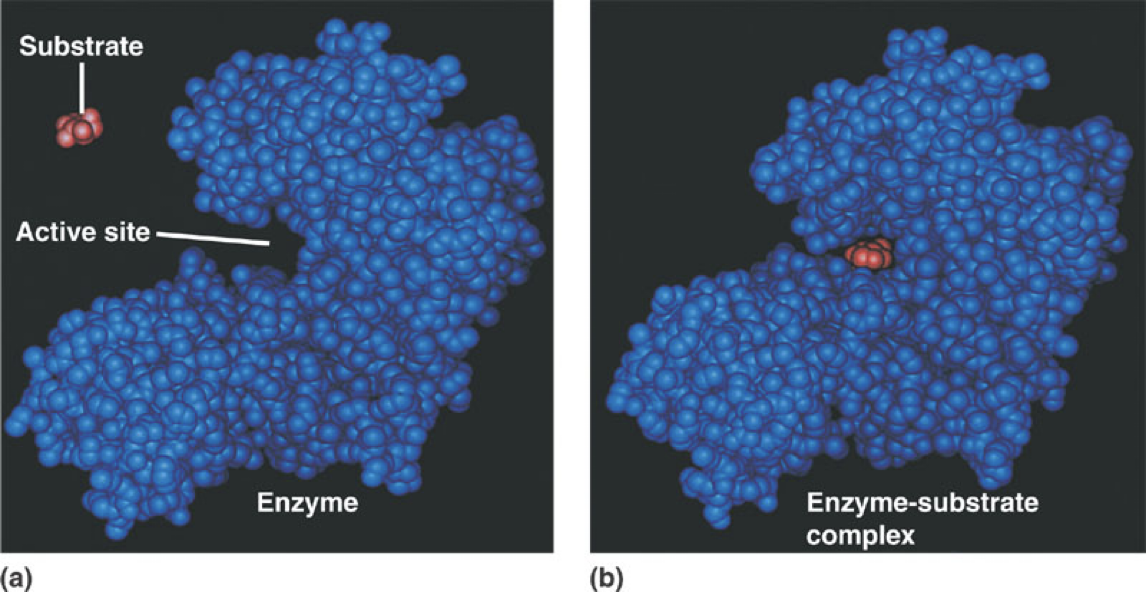
Induced Fit Model: Enzyme shape changes upon substrate binding to improve catalytic activity of the enzyme
-Catalytic Efficiency and the Active Site-
Substrates are held in close proximity and proper orientation
Formation of covalent intermediates
Active site contains functional groups that temporarily donate H+s or e-s
Binding of a substrate to a enzyme induces the strain on the substrate molecules and makes it more reactive
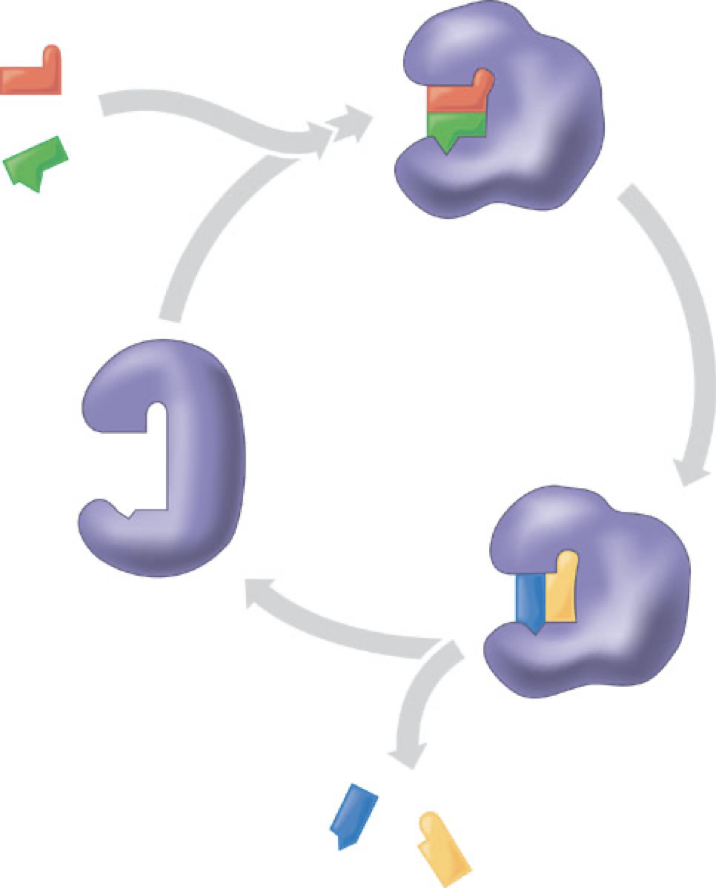
The active site lower the activation energy
Acts as a template for the reaction
It stresses the substrate
Stabilizes the transition state
Participates in catalytic reaction
Slides 75-95: Factors Influencing Enzyme Activity
A proteins shape is essential to its function
A denatured protein loses its 3D shape, and becomes an unfolded chain of amino acids
A denatured protein will not function properly
During denaturation the ionic, H bonds and disulfide bonds that hold the protein in its 3D shape are broken, and the enzymes unfolds
The amino acids are still connect in a chain by peptide bonds
-What Causes Protein Denaturation-
Large change in pH
Acidic or basic
High temperature
Denaturation is usually permanent
When an enzyme reaches past its optimal temperature or pH the enzyme are permanently inactivated when they are denatured
-Small Changes in pH (<1 pH unit)
Will change the charge on the substrate and the charge on the enzyme (active site)
The substrate may no longer be able to bind as well, so the enzyme will be less active
pH optimum: The pH at which the enzyme performs the reaction on the maximum rate
The enzyme becomes denatured above and below the pH optimum, so its is less active
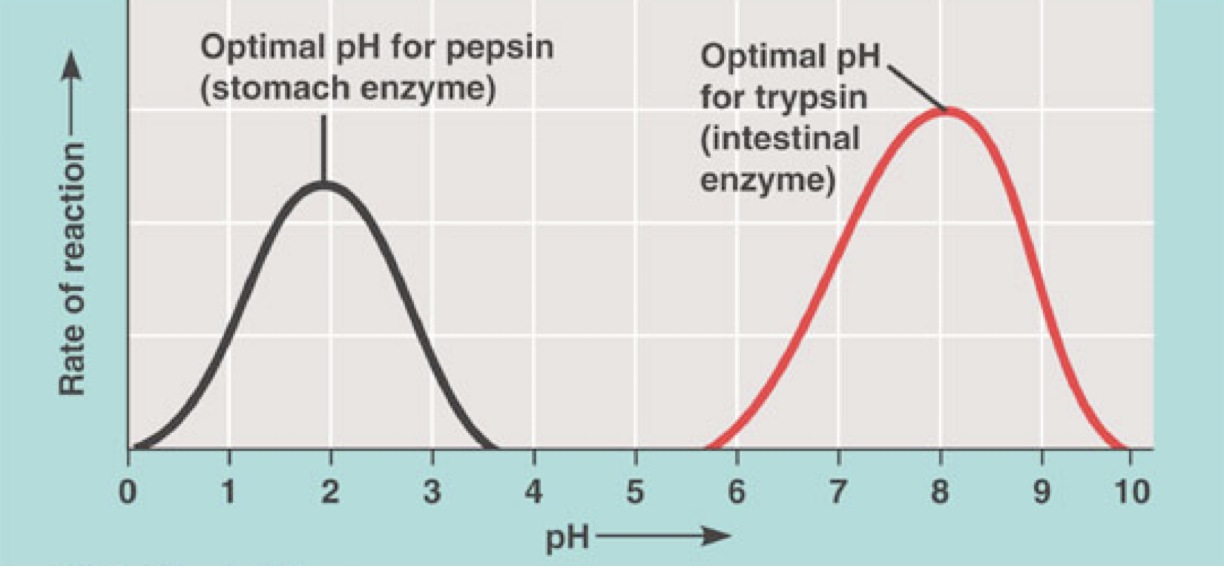
Most enzymes have a pH optimum that matches the pH of the environment the enzyme is designed to work in
-Large changes in pH (>1 pH unit)
Will break the ionic and H bonds that hold protein in its 3D shape
Enzymes may partially unfold and become less active
Enzymes may completely unfold (denature) and become totally inactive
-Temperature and Enzyme Activity-
Many enzymes have a temperature optimum that is equal to the body temperature of the organism they are from
Slides 96-101: Kinetic Energy and Reaction Rates
Kinetic Energy: energy associated with movement
Molecules are always moving: they have kinetic energy
More kinetic energy → more collisions between molecules → Faster reaction rate
The colder the temperature the mess kinetic energy a molecule has
Which result in fewer collisions between molecules → lowers reaction rate
If there are fewer collision between reactant molecules at a low temperature the reaction rate will be slower
Slides 97-106: Additional Factors for Maximal Enzyme Activity
Prosthetic Group: small organic molecules permanently bound to a enzyme
Coenzymes: small organic molecule (vitamin derivaties), bind temporarily to enzymes
Cofactors or Metal Ions: ex. Fe, Mg
-Function-
May be required for proper 3D shape of protein
May be directly involved in chemical reaction
Coenzymes may change the shape of the active site
Coenzymes may make a temporary bond between 2 substrates
Inhibitor: molecule or ion that binds to a enzyme and decreases its activity
Competitive inhibitor binds to the active site of a enzyme
The inhibitor must resemble the substrate in order to bind to the active site
If the inhibitor binds to the active site the substrate can’t bind, so no reaction will occur
The more inhibitor is added, the more the reaction is inhibited, there are mnay enzyme molecules in the cell
The more inhibitor binds to the active site the lower the reaction rate
The inhibitor binds to the active site, so it must have a similar structure to the normal substrate
Noncompetitive Inhibitors: Bind to allosteric site noncovalently, altering enzyme shape
Some proteins have more than one subunit
Catalytic subunit: contains the active site and performs reactions
Regulatory subunit: contains the allosteric site and regulates enzyme activity
Inhibitor binds to allosteric site on regulatory subunit
Changes the shape of regulatory subunit and the catalytic subunit
Enzyme activity decreases
Page 122-138: Allosteric Regulation
Catalytic Subunit: Binds substrate at the active site, performs enzyme reaction
Regulatory subunit: binds an effector at the allosteric site, controls activity of enzyme
Effector: small organic molecule that controls the activity of the enzyme
The effector binds to the allosteric site
Active form: form of the enzyme that performs its function (ON)
Inactive form: nonfunctional form of enzyme (OFF)
Effector controls activity of enzyme
Regulatory molecules / effectors binds to regulatory subunit and causes it to change shape
A change in shape of the regulatory subunit causes catalytic subunit to change its shape
Catalytic subunit no in inactive form (OFF)
Regulatory subunit may still be bound to catalytic subunit but both change its shape
An inhibitor binds to the regulatory subunit
inhibitor = effector
Inhibitor binding causes regulatory subunit to change shape
Change of shape of regulatory subunit causes catalytic subunit to change shape, from active form to inactive form
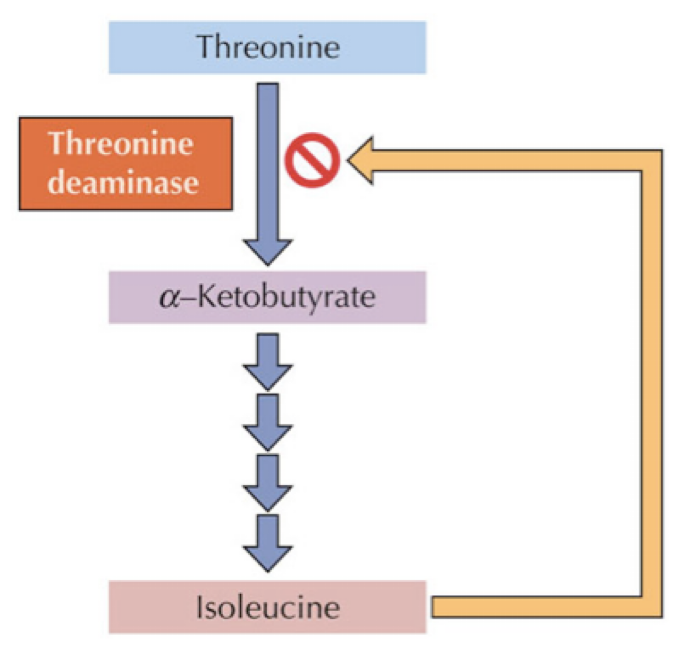
Accumulation of product of a metabolic pathway inhibits one of the enzymes at the beginning of the pathway
Feedback inhibition is a kind of allosteric inhibition
Inhibitor = product of pathway
Inhibitor binds to allosteric site on regulatory subunit of enzyme 1
Regulatory subunit changes shape
Catalytic subunit changes shape → inactive form
When a product made at the end of a pathway binds to one of the enzyme near the beginning of the pathway, and inactivates the enzyme
Product binds to enzyme 1 at the allosteric site and changes the enzymes shape
Enzyme 1 : Active → inactive (OFF!)
Allosteric regulation is a kind of noncompetitive inhibition
it is used to regulate enzymes in pathways
Feedback inhibition is a kind of allosteric inhibitor
Allosteric inhibitor: binding of the effector makes enzyme less active
Active enzyme → inactive (OFF)
Allosteric activator: binding of the effector makes enzyme more active
Inactive enzyme → Active (ON)
Page 139: Summary of Allosteric Regulation
Allosteric regulation is crucial for maintaining metabolic balance
Involves both activation and inhibition mechanisms