Genotyping of ALDH2 gene by allele-specific PCR
Aldehyde dehydrogenase
reaction
CH3CH2OH → (by alcohol dehydrogenase) CH3CHO (acetaldehyde) → (by ALDH2) CH3COO- (acetate)
deficiency of ALDH2
acetaldehyde cannot be converted to acetate
→ accumulation of acetaldehyde
→ alcohol flush response
SNP in ALDH2 gene
rs671: SNP (G to A) at codon 504
E504K: amino acid translated changes from Glu to Lys
Allele-specific PCR
General SNP genotyping
to determine the SNP allele at each SNP marker in each sample
methods includes
allele-specific PCR
PCR-RFLP
DNA sequencing
microarray
primer extension
PCR——polymerase chain reaction
purpose: amplify a specific DNA fragment from a complex mixture of DNA molecule
procedures
strand separation (95°C)
heat denaturation of template DNA
primer annealing (58°C)
forward primer: same as the top strand
reverse primer: reverse complement of the top strand
strand elongation (72°C)
DNA synthesis always proceeds from 5’ to 3’ end of the growing strand
DNA polymerase adds dNTPs to the free 3’-OH end of the primer
Mg2+ is required as cofactor
★ PCR products contain primer sequences
General info of Allele-specific PCR
also known as amplification refractory mutation system (ARMS)
use allele-specific primer for PCR
presence or absence of PCR product indicate the presence or absence of the target allele
mismatch at 3’-OH terminal of primer→ drastically reduce amplification efficiency to differentiate different alleles
require DNA polymerase without 3’ to 5’ exonuclease activity
proofreading activity of DNA polymerase remove the mismatched base at the 3’ end of the growing DNA → cannot differentiate the alleles
Components require for the reaction
buffer
template DNA
primers
dNTPs
thermostable DNA polymerase
divalent cation: Mg2+ for catalytic activity of DNA polymerase
may cause min incorporation of bases if Mn2+ is used
Primer design
avoid formation of primer dimers: no complementary sequences on 3’ ends of both primers
primer dimers= primers anneal to each other
this leads to fewer primers available for annealing
avoid inverted repeat in primers
→ primer self-annealing and no primer extension
18-30 nucleotides long
PCR annealing temperature
melting temperature of primer= (no. of A + T)*2°C + (no. of G + C)*4°C [for short oligonucleotides up to 20 bases]
melting temperature of forward and reverse primer should not differ by > 5°C
set the temperature of annealing a few degrees below the melting temperature
too high: primer may not anneal → poor amplification
too low: tolerate mismatch between primer and template → non-specific amplification
additional deliberate mismatches at the penultimate base of the primer
increase discrimination between alleles
Multiplex PCR
amplify more than one gene in a single PCR reaction
serving as positive control: to show the PCR is functioning → the absence of PCR product in allele-specific PCR is not due to
failure of PCR or
absence of genomic DNA template
Agarose gel electrophoresis
Principle
for analysis of DNA and purification of DNA fragments for cloning
usually run submerged in running buffer
DNA or RNA are -ve charged: migrate towards +ve electrode when in electric field
DNA or RNA molecules have a constant charge-to-mass ratio
migrate according to their size
smaller molecules migrate faster
mobility of DNA is inversely proportional to the log of the number of pairs in DNA
Procedures
add DNA samples and DNA ladder (size marker) onto agarose gel lane
DNA bands are separated by size from negative electrode to positive electrode
add dye to bind to DNA
expose DNA bands on film: bands become visible Unser UV light
Equipment required
agarose
a liner polysaccharide made up of the basic repair unit agarobiose
solution in hot water: gel is formed via the corsslinking of agarose polymer chain by interchain hydrogen bond when cools down
size of pores: 50-200 nm (depends on concentration of agarose)
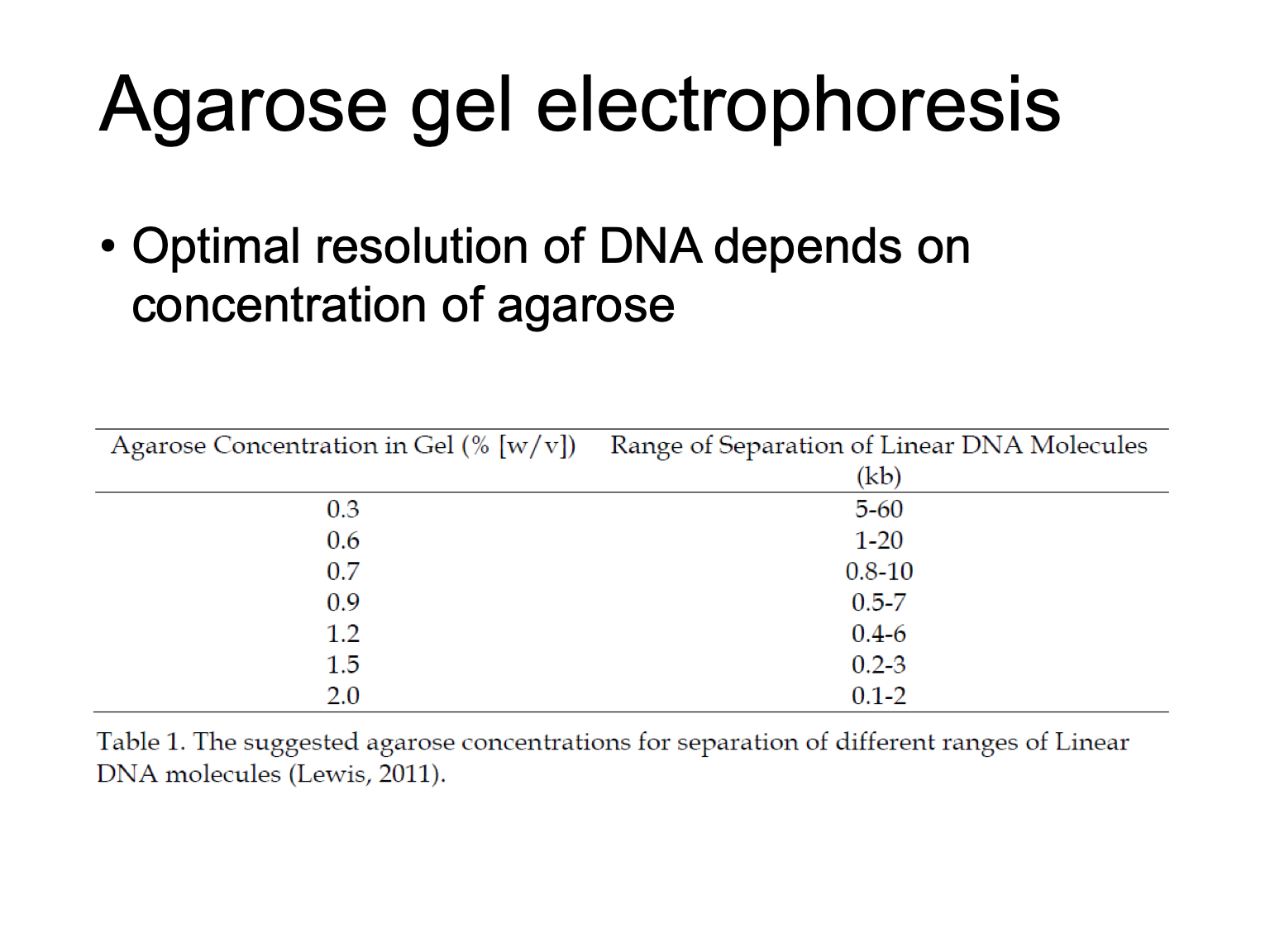
As the agarose concentration increases, the average diameter of the pore decreases
optimal resolution of DNA depends on concentration of agarose
electrophoresis chamber and power supply
gel casting tray
sample combs
around which molten medium is poured to form wells in gel
electrophoresis buffer
TAE
lower buffering capacity
may need recirculation of buffer
preferred if extraction of DNA from gel is needed or separating large DNA molecules
TBE
higher buffering capacity
used DNA recovery is not needed
slower DNA migration but sharper bands are produced
pH 8.3
usually prepared as 10X or 5X and dilute to 1X working concentration when use
must use the same buffer for gel preparation and electrophoresis
loading dye or buffer
contains glycerol, Ficoll or sucrose to provide sample density for loading
6X or 10X concentrated form
contains tracking dyes to monitor the progress of electrophoresis
xylene cyanol co-migrates with DNA fragments ~ 4kb (slowest)
bromophonel blue co-migrates with DNA fragments ~ 0.4 kb
orange G co-migrates with DNA fragments ~ 50bp (fastest)
DNA intercalating dye
include in gel or/and buffer to visualise DNA
stain gel with DNA binding dye after electrophoresis to visualise DNA
higher affinity to double stranded DNA
Radiation at 302 nm and 366 nm is absorbed by dye → re-emits as fluorescence at 590 nm (red-orange)
examples
ethidium bromide
GelRed
other dyes such as SYBR Gold, SYBR Safe, SYBR Green and methylene blue
UV transilluminator
visualise DNA at 302 or 366 nm
photography with gel documentation system
determine the approx. length of DNA molecule by comparing their migration to that of standards
Factors affecting agarose gel electrophoresis of DNA
DNA size (inversely proportional to the lot of the number of base pair)
concentration of agarose
different concentration for different size range
voltage
Migration of DNA on agarose gel roughly proportional to voltage x hour
use lower voltage for running larger size of DNA fragments
DNA topological form
nicked circular: slower
linear (size marker is designed for linear DNA fragments only)
superhelical: faster