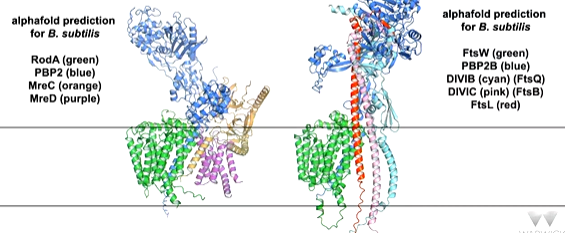
lecture 12: bacterial cell wall peptidoglycan biosynthesis 2
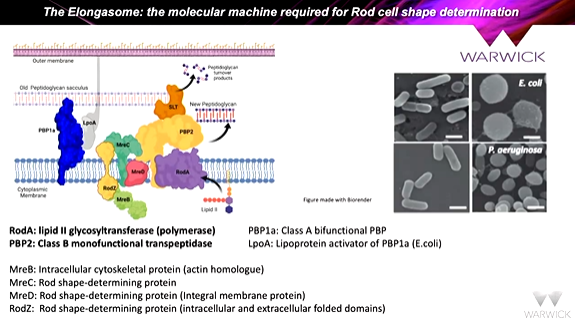
the proteins at the bottom along with PBP2 forms the elongosome. Inhibition of PBP2 makes the bacteria round. It was shown that making a fusion protein of RodA and PBP2 mirrored what was happening at the genome level as the genes are in the same operon so are transcribed and translated at the same time to form a complex. The idea of fusing teh proteins together is a good one and expressed in e coli and extracted from membranes using detergents. This is stabilized using a membrane scaffold protein - msp nanodisc - which forms a belt around a membrane protein to keep it stable in solution. It resulted in individual rodA-PBP particles. This means that the EM data can be collected and group the images together to show class averages. The class averages of orientations were taken and put together to give a structure.
It was shown in the cryo-em structure that PBP2 has a single TM helix extends into the 10 membrane helices of RodA. This is a tight complex. This intercalation open rodA like a flower and activate sit as a glycosyl transferase. This is a functionally dependent complex!!.
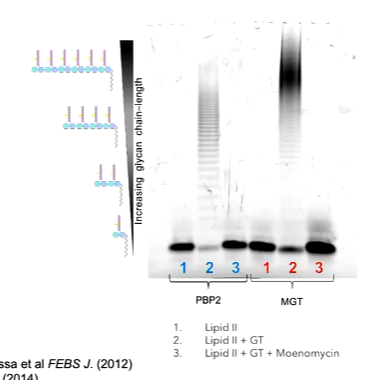
If you take all the sequences of SEDS proteins from 20 bacterial pathogens and sequence align them with a logo plot to show the frequency of the appearance of individual amino acids, the important residues can be seen. There are some which are important such as tryptophan and others such as a sequence of proteins and tyrosine and glycine. We are not entirely sure why these are conserved. W102,R210 and D262 are focused on here. To interrogate the function of a residue in a protein can be discovered through site directed mutagenesis. This means that you can change the identity of the amino acid and probe what it does. You also need a way to test what happens to the protean - Tris-Tricine polyacrylamide gel assay by making lipid II. This is made fluorescent by appending a dysyl group to make Lipid Ii glow yellow. in a gel based assay, you can see lipid II as it runs quickly down the gel but any polymers run up the gel.
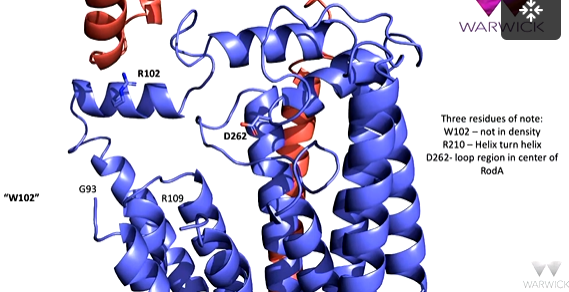
The three residues of interest are in RodA, R is in the helix turn helix motif, D262 is in a loop region in the centre of RodA and is adjacent to two cavities. W102 is not able to be seen in the density in the CroEM structure which suggests that it is flexible.
Aspartate 262 is the active site residue and is responsible for catalysing the reaction. Mutation to Ala kills it stone dead and so Lipid II cannot be polymerised. There are other residues that are also important to this but the main one is D. UDP is captured in the protein. D262 as an acid abstracts a protein at the 4th position of the incoming lipid II to make an oxygen nucleophilic. This can attack the position 1 of the next Lipid II to form the 1,4 beta glycosidic bond. This is exactly analogous to the mechanism for the possessive glycosyl transferase reaction
R on the helix turn helix motif is required to keep the UDP in the right place at the right time. The helix turn helix motif has high mobility so can move and moves in and out of the protein to sweep in or out UDP. When this is mutated to Ala, it is killed
The Tryptophan is mutated to the Ala, the enzyme is killed but if it is mutated to Phenylalanine, it is still active. This is because the aromatic ring is still retained om the side chain. The W on the mobile loop guides through a pi-pi stacking interaction on the glycan strands out of the active site. In other enzymes with a sugar polymer forming activity, there is a W in the same position. It cannot be seen in the density, so this is not fully understood.
There are two large cavities in RodA where lipid II fits and this is the equivalent to the acceptor and donor sites. the active site aspartate is between these two. The three residues are important for overall function of the protein
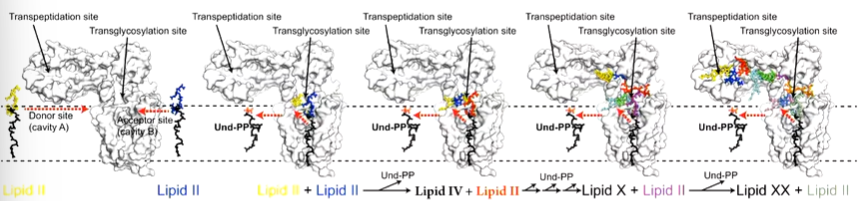
two lipid Iis approach the cavities with one going to the donor site and one to the acceptor site. The reaction of the glycosyltransferase proceeds and UDP is released to form a lipid 4 molecule. Another lipid II comes in to extend the polymer. RodA opens and closes to allow the entry of lipid II.
you find in other enzyme that utilize UDP carrier lipid all have a highly conserved R on the helix. In some of these structures, you see the whole UDP through the protein. Using alphafold, if you compare the experimental structure with the alphafold predicted structure, you see the alphafold structure predicts the enzyme extends with a conformational change between the experimental state with no ligands and the predicted active state which is extended and can reach into the peptidoglycan layer to introduce more polymer. There are other proteins associated with this