ch15 ; the chromosomal basis of inheritance
Mendel’s “hereditary factors” were abstract when proposed, but know we know that these factors are genes and are located on chromosomes. You can see this location by tagging isolated chromosomes with fluorescent dye that highlights the gene.
Cytologists worked out the process of mitosis in 1875 and meiosis in 1890s using improved techniques of microscopy.
Biologists began to see parallels between the behavior of Mendel’s proposed hereditary factors and chromosomes.
Around 1902, Sutton and Boveri and others independently noted these parallels and began to develop the chromosome theory of inheritance: that genes are found at specific locations on chromosomes.
Morgan showed that Mendelian inheritance has its physical basis in the behaviour of chromosomes: Scientific Inquiry
The first solid evidence associated a specific gene with a specific chromosome came in the early 1900s from the work of Thomas Hunt Morgan. In one experiment, Morgan mated male flies with white eyes (mutant) with female flies with red eyes (wild type)
The F1 generation all had red eyes
The F2 generation showed a 3:1 red to white eye ratio, but only males had white eyes.
Morgan reasoned that the white eyed mutant allele must be located on the x chromosome
this is because the males were showing traits of white eye (they only need 1 x to show the trait whereas females would need 2 recessive x)
Morgan’s findings supported the chromosome theory of inheritance
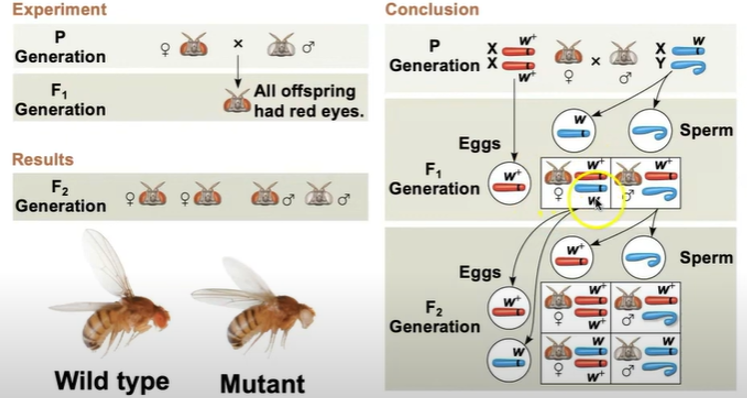
The wild type allele is W+ and the mutant allele is w. when a female produces gametes, there’s a 50/50 chance on which gene she will give (it’s one or the other.) and same with males, 50/50 whether they give an x or a y. So the F1 generation will have a wild type and a mutant type (she’s a carrier) and the male will get only the wild type and the y chromosome (males cannot receive X from the father). The F2 generation would have 1 male that receives a mutant chromosome from mom (so it has mutant phenotype) and 2 females (1 carrier, one wild type) and 1 male that is wild type.
Sex Linked Genes Exhibit Unique Patterns of Inheritance: The Chromosomal Basis of Gender.
In humans and some other animals, there is a chromosomal basis of sex determination. There are two varieties of sex chromosomes: a larger x chromosome and a smaller y chromosome
Y chromosomes usually only have a couple 100 genes, and they usually just direct male development
Only the ends of the y chromosome have regions that are homologous with corresponding regions of the x chromosome
The SRY gene on the y chromosome codes for a protein that directs the development of male anatomical features
Females are XX, males are XY. Each ovum contains an X chromosome, while a sperm may contain either an X or a Y chromosome. A gene that is located on either sex chromosome is called a sex linked gene.
genes on the y chromosome are y linked genes (rare, there are only a few of these)
genes on the x chromosome are called x linked genes.
Inheritance of X-Linked Genes
X chromosomes have genes for many characters unrelated to sex, whereas the y chromosome mainly encodes genes related to sex determination. X-linked genes follow specific patterns of inheritance. For a recessive x-linked trait to be expressed:
Female needs two copies of the allele (homozygous)
Male needs 1 copy of the allele (hemizygous)
X-linked recessive disorders (color blindness, hemophilia, and Duchenne muscular dystrophy) are much more common in males than in females (since a male only needs one recessive X to show the trait).
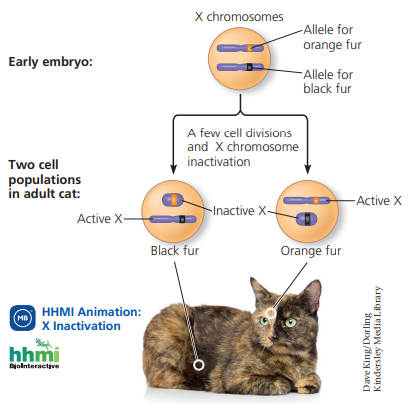
X-inactivation in female mammals
In mammalian females, one of each of the two x chromosomes in each cell is randomly inactivated during embryonic development (bc we don’t need two cells active for the same function). The inactive X condenses into a Barr body. In a female is heterozygous for a particular gene located on the x chromosome, she will be a mosaic for that character
E.g., tortoiseshell cats
Linked genes tend to be inherited together because they are located near each other on the same chromosome
Each chromosome has hundreds or thousands of genes (except the y-chromosome). Genes that are located on the same chromosome tend to be inherited together are called linked genes.
We can figure out how close to genes are located on the chromosome (if they are “linked”) by checking how often they are inherited together. If two characters show up more in pairs, they’re likely linked.
Recombination of Unlinked Genes: Independent Assortment of Chromosomes
Offspring with a phenotype matching one of the parental (P) phenotypes are called parental types. Offspring with non-parental phenotypes (new combinations of traits) are called recombinant types or recombinants.
50% frequency is observed for nonlinked genes, with 50% of offspring being parental types, and 50% of offspring being recombinants.
Recombination of Linked Genes: Crossing Over
Morgan observed that although some genes are linked, non-parental allele combinations are still produced. He proposed that some process must occasionally break the physical connection between genes on the same chromosome. That mechanism was the crossing over of homologous chromosomes.
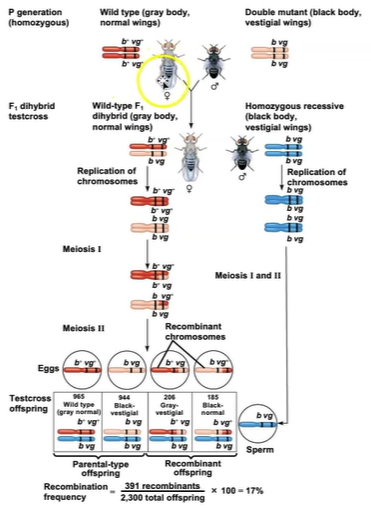
Calculating recombination frequency:
Calculate the number of recombinant offspring divided by the total number of offspring.
The lower the number of recombination frequency, the closer together the alleles are on the chromosome.
Once the frequency hits 50%, the alleles are likely on different chromosomes altogether.
New Combinations of Alleles: Variation for Natural Selection
How does this cause variation in species?
1) Recombinant chromosomes bring alleles together in new combinations in gametes.
2) Gametes random fertilization increases even further the number of variant combinations that can be produced.
Mapping the Distance Between Genes using Recombination Data: Scientific Inquiry
Alfred Sturtevant, one of Morgan’s students, constructed a genetic map, an ordered list of the genetic loci along a particular chromosome. Sturtevant predicted that the farther apart two genes are, the higher the probability that a crossover will occur between them and therefore the higher the recombination frequency. A linkage map is a genetic map of a chromosome based on recombination frequencies. Distances between genes can be expressed as map units; one map unit = 1% recombination frequency. Map units indicate relative distance and order, not precise locations of genes. Genes that are far apart on the same chromosome can have a recombination frequency near 50%
These genes are physically linked, but genetically unlinked, and behave as if found on different chromosomes.
Sturtevant used recombination frequencies to make linkage maps of fruit fly genes. He and his colleagues found that the genes clustered into four groups of linked genes (linkage groups)
The linkage maps, combined with the fact that there are four chromosomes in Drosophilia, provided additional evidence that genes are located on chromosomes.

The recombination frequency here is 17%, so on a genetic map it would be 17 map units, which is how far apart b and vg are. Cn is another gene, and we can calculate that there a 9 map unit distance between cn and b, and 9.5 map units between cn and vg.
Abnormal Chromosome Number
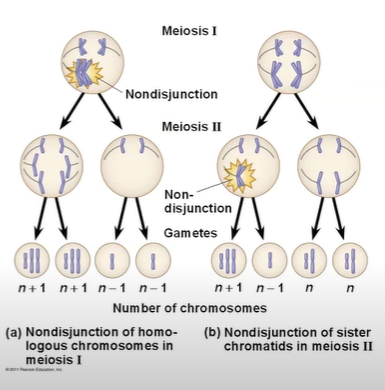
In nondisjunction, pairs of homologous chromosomes do not separate normally during meiosis. As a result, one gamete receives two of the same type of chromosome, and another gamete receives no copy.
it’s usually more detrimental when it occurs in meiosis one, because then every single gamete has an abnormal amount of chromosomes. When it happens in meiosis ii, only 2 gametes produce an abnormal amount of chromosomes.
Anueploidy results from the fertilization of gametes in which nondisjunction occurred. Offspring with this condition have an abnormal number of a particular chromosome
A monosomic zygote has only one copy of a particular chromosome
A trisomic zygote has three copies of a particular chromosome
trisomy 21 - Down syndrome
Triploidy (3n): 3 sets of chromosomes
Tetraploidy (4n): 4 sets of chromosomes
Down Syndrome (trisomy 21)
Down syndrome is an aneuploid condition that results from three copies of chromosome 21. It affects about one out of every 700 children born in the united states. The frequency of Down syndrome increases with the age of the mother, a correlation that has not yet been explained.
Aneuploidy of Sex Chromosomes
Nondisjunction of sex chromosomes produce a variety of aneuploid conditions
Klinefelter syndrome is the result of an extra chromosome in a male, producing XXY individuals
Monosomy X, also called Turner Syndrome, produces X0 females, who are sterile; it is the only known viable monosomy in humans.
Alterations of Chromosome Structure
Breakage of a chromosome can lead to four types of changes in chromosome structure.
Deletion removes a chromosomal segment
deletion in chromosome 5 results in the syndrome “cri du chat”
this results in severe intellectual disability and a catlike cry; individuals usually die in infancy or early childhood.
Duplication repeats a segment
Inversion reverses orientation of a segment within a chromosome
Translocation moves a segment from one chromosome to another
certain cancers, like chronic myelogenous leukemia (CML), are caused by translocations of chromosomes.
Some Inheritance Patterns are Exceptions to Standard Mendelian Inheritance
There are two normally occurring exceptions to Mendelian genetics
Genes located in the nucleus
For a few mammalian traits, the phenotype depends on which parent passed along the alleles for those traits
This variation in phenotype is called genomic imprinting
Genomic imprinting involves the silencing of certain genes depending on which parent passes them on.
It seems that imprinting is the result of the methylation (adding of CH3) of cysteine nucleotides
Methyl groups are involved in gene expression, they can control whether a certain protein is made. Methylation closes off the chromosomes, stopping it from continuing to be made.
Genomic imprinting may affect only a small fraction of mammalian genes
Most imprinted genes are critical for embryonic development.
Genes located outside the nucleus
Extra-nuclear genes (cytoplasmic genes) are found in organelles in the cytoplasm
Mitochondria, chloroplasts, and other plant plastids carry small circular DNA molecules
Extra-nuclear genes are inherited maternally because the zygote’s cytoplasm comes from the egg
Mitochondria are solely inherited from the mother
The first evidence of extra-nuclear genes came from studies on the inheritance of yellow or white patches on leaves of an otherwise green plant
Some defects in mitochondrial genes prevent cells from making enough ATP and result in diseases that affect the muscular and nervous systems
E.g., mitochondrial myopathy and Leber’s hereditary optic neuropathy
It may be possible to avoid passing along mitochondrial disorders
The chromosomes from the egg of an affected mother could be transferred to an egg of a healthy donor, generating a “two-mother” egg. This egg could then be fertilized by sperm from the prospective father and transplanted to the womb of the prospective mother
In both cases, the sex of the parent contributing an allele is a factor in the pattern of inheritance.
STUDY REVIEW QUESTIONS:
1. Predict the ratio for different offspring based on parental genotypes
To predict the ratio for different offspring based on parental genotypes, you can use probability calculations.
The product rule: the probability of two or more independent events occurring is calculated by multiplying the probability of each event. Used for unlinked genes. If X “and” Y occur.
What’s the probability that the offspring of two heterozygous parents will be homozygous recessive?
They need a recessive gene from mom AND a recessive gene from dad.
Probability of receiving a recessive gene from mom? ½
Probability of receiving a recessive gene from dad? ½
½ x ½ = ¼
The probability of the offspring being homozygous recessive is ¼, or 25%
The sum rule: the probability of two or more mutually exclusive events is calculated by adding the probability of event X and event Y. If X “or” Y must occur.
What is the probability that the offspring of a heterozygous parents will NOT be homozygous recessive?
They would either be heterozygous OR homozygous dominant.
Probability of being heterozygous: ½
probability of receiving a recessive from mom and a dominant from dad":
½ x ½ = ¼
probability of receiving a recessive from dad and a dominant from mom:
½ x ½ = ¼
¼ + ¼ = ½
Probability of being homozygous dominant: ¼
probability of receiving dominant from mom AND a dominant from dad:
½ x ½ = ¼
Probability of being heterozygous OR homozygous dominant?
½ + ¼ = ¾
The probability of the offspring not being homozygous recessive is ¾.
(Alternatively, you can do P(all events occurring) - P(homozygous recessive), which would be 1 - ¼ = ¾)
For dihybrids and beyond, you can use the above rules to calculate the probability.
In the offspring between AaBbCc x AabbCc, what is the probability that the offspring will show all recessive traits?
Offspring must be aa “and” bb “and” cc, so we use multiplication rule.
probability of receiving a from both parents: ¼
probability of receiving b from both parents: ½
(one parent always gives b= 1, other parent gives a b ½, so 1 × ½)
probability of receiving c from both parents: ¼
therefore, the probability of receiving all recessive traits: ¼ x ½ x ¼ = 1/16
2. Identify an inheritance pattern from a pedigree
inheritance pattern | identifiers | examples |
|---|---|---|
autosomal recessive |
|
|
autosomal dominant |
|
|
y-linked |
| |
x-linked recessive |
|
|
x-linked dominant |
|
|
mitochondrial |
|
|
3. Calculate the risk of a certain individual in a pedigree being a carrier for a gene
We can calculate the risk of an individual being a carrier by identifying their genotype. To do so:
identify the inheritance pattern of the gene
look at the genetic history of the individual
by identifying their parents genotypes based on the inheritance pattern of the trait, you can predict the likelihood of the offspring being a carrier or not.
If my maternal grandfather had colorblindness (x-linked recessive) and my father has colorblindness, what is the likelihood that I, a female, am a carrier?
Because my father has colourblindness, I am 100% a carrier. This is because my father’s genotype is xY, and every daughter receives an x chromosome from her father. Therefore, I do have the recessive gene for colourblindness in the x chromosome I inherited from my father.
What is the likelihood that my child will be a carrier if I marry a non-colourblind man?
Since my genotype is xx, and my husband’s is xy, there is a ½ chance my son will present colorblindness, a 0% chance that my daughters will be colourblind, a 0% chance that my sons will be carriers (males cannot be carriers in colourblindness), and a 50% chance my daughter will be a carrier (she can either be xx or xx).
4. Explain the outcome of X inactivation
X inactivation occurs because females do not need both x chromosomes, so in each cell one of the x chromosome is randomly inactivated. When the x chromosome has alleles that differ from one another (she is heterozygous for the gene), the x that remains active shows the trait. In tortoiseshell cats, this shows up as random cells being either coding orange or black fur, showing up as a mosaic.
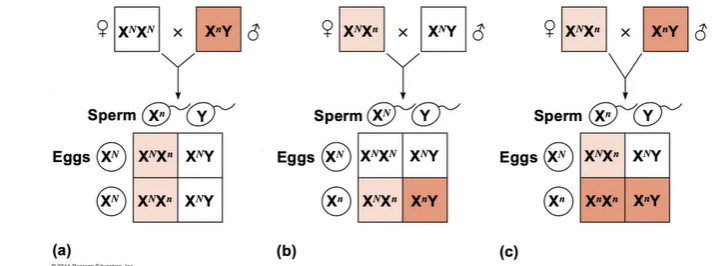
5. Predict which parent nondisjunction occurred in, based on an individual’s karyotype or family tree genetic disease history.
X0 : can come from mom or dad
XXX: can come from mom OR dad
XXY: can come from mom OR dad
XYY: can come from dad only
You can use the phenotype or genotype of the offspring and family to determine which parent it came from.
A boy with CGH is born with Klinefelter syndrome. His mother does not have CGH, but his father also has CGH. In which parent did nondisjunction occur?
In Klinefelter syndrome (XXY), the extra x could’ve come from the dad or the mom.
Since this is an x-linked disorder, we know that the father must’ve given both x AND y chromosomes to the son, since the mother had no way of transmitting this disorder. This means that nondisjunction occurred in the father, not the mother.