The Genetic Code and Transcription
Vocabulary:
Central Dogma of Molecular Biology: Describes the unidirectional flow of genetic information from DNA to RNA to protein, transcription copies DNA into an mRNA sequence, translated in ribosomes using tRNA into an amino acid sequence
Messenger RNA (mRNA): The complement of a template strand, acts as a messenger molecule being transported out of the nucleus, will associate with ribosomes where it is decoded and translated into a protein
Triplet code: “words” for genetic code, consists of 3 ribonucleotide letters, 64 unique arrangements that only specify 20 amino acids
Codon: Consists of one triplet code, has 3 ribonucleotides
Unambiguity: A property of genetic code in translation, each codon only corresponds to one amino acid
Degeneracy: A property of genetic code in translation, a given amino acid can correspond to multiple codons
Commaless: A property of translation, once there is a start signal codons are read with no break, until there is a stop signal
Nonoverlapping: A property of translation: a single ribonucleotide within mRNA is only part of one triplet
Colinear: A property of genetic code in translation, the sequence of codons determines the order of the amino acids in the encoded protein
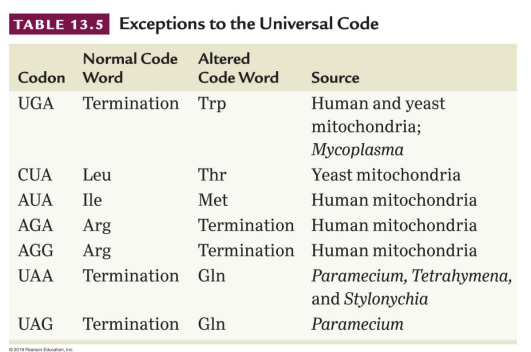
Universal: A property with only one exception in translation, a single coding dictionary is used by all organisms including viruses, prokaryotes, archaea, and eukaryotes
Methionine (MET): An amino acid encoded by the AUG triplet, can initiate a polypeptide chain
Termination signal: A triplet that instead of encoding an amino acid will stop polypeptide synthesis, UAA UAG of UGA
Inosine: A base in tRNA that can match with A, U, or C
Reading frame: Where mRNA will start reading 5’→3’ mRNA, based on a contiguous sequence of nucleotides, though disrupted by frameshift mutation will be reestablished with 3 insertions/deletions
Frameshift mutation: A mutation that will change the reading frame, caused by an insertion or deletion of a nucleotide
Sydney Brenner: Coined the term ‘codon’ in the 1960s, theorized mathematically that it was a triplet
Nirenberg and Matthaei: In 1961 deciphered the first codon UUU, encoding phenylalanine and showed that RNA controlled the production of proteins. Extracted the cellular machinery from E. coli responsible for protein production and demonstrated that proteins could be made when to intact living cell was present, used cell-free protein synthesis to link specific coding sequences to specific amino acids
Cell-Free Protein Synthesis: Done in cell lysate, which had all essential factors for protein synthesis without organelles or a cell membrane, uses the enzyme polynucleotide phosphorylase to synthesize RNA templates for mRNA, generates random RNA
RNA Homopolymer codes: RNA molecules that only contained a single type of nucleotide
tRNA: A kind of RNA used in translation, contains an anti-codon and is attached to an amino acid
Anti-codon: A complementary sequence to mRNA that corresponds to a single amino acid
Nirenberg and Leder: Looked into how specific sequence assignment for triplet codons was made possible by triplet binding assay, observed that ribosomes could bind sequences as short as 3 ribonucleotides. Radioactively labeled amino acids and added them to a cell-free system, enzymes in the lysate attached the radioactive amino acids to tRNAs, used knowledge of codon composition to narrow down which amino acid was each specific triplet. Concluded code was degenerate and unambiguous
Triplet binding assay: The experiment that led to the sequencing of every amino acid. Knew the compositions of each polyribonucleotide, and could synthesize tRNA with unique sequences. Radioactively labelled the amino acids, and filtered them by size which excluded tRNAs that were bound to codons
Wobble hypothesis: The initial two ribonucleotides of the triplet codes are often more critical than the third. This allows for degeneracy using the third positioned ribonucleotide in tRNA, it is less spatially constrained and doesn’t need to adhere as strictly to established base-pairing rules
N-formylmethionine (fmet): A modified form of methionine found in bacteria
Nonsense mutations: A mutation that can produce a stop codon internally prematurely, terminating translation and producing only a partial polypeptide
Mitochondrial DNA (mtDNA): Has many exception to universal genetic code, UGA typically specifies termination but encodes tryptophan in yeast and humans. AUA normally encodes isoleucine but encodes internal insertion of methionine
Overlapping genes: When a single mRNA strand may have multiple initiation points, which creates different reading frames and may specify more than one polypeptide. This is common in viruses
Open reading frame (ORF): Happens with overlapping genes, a series of triplet codons will specify amino acids to make a polypeptide and initiation at different AUG positions within the frame will results in distinct polypeptides
Template strand: The strand in DNA that is actually transcribed into mRNA in the 5’→3’ direction
Coding strand: The strand of DNA that is not transcribed
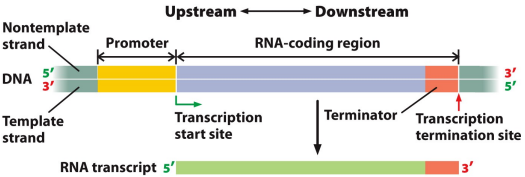
Promoter: A regulatory sequence upstream of the 5’ end where RNA is transcribed, part of DNA, non-coding but where the transcription machinery binds, identifies where a gene starts and which strand to use as the template, giving it orientation. Not transcribed itself
RNA-coding region: The part of DNA that is transcribed into mRNA, will likely contain protein coding and protein non coding sequences
Terminator: Part of DNA, ends transcription, consists of a regulatory sequence, part of the mRNA transcript created
RNA Polymerase: An enzyme that directs the synthesis of RNA from a DNA template, doesn’t use a primer and its nucleotides contain ribose not deoxyribose
Bacterial RNA Polymerase: Contains 5 subunits, 2 alpha, and one of each beta, beta’, and omega. The holoenzyme contains an additional sigma factor, which can bind to a promoter and initiate transcription. Different sigma subunits recognize different subsets of promoters, this mechanism is used to regulate gene expression under varying environmental conditions like heat/starvation. The sigma subunit dissociates while elongation proceeds with just the core enzyme (50/sec at 310 K)
Initiation: A mechanism of transcription, it is the assembly of apparatus at the promoter and the start of RNA synthesis
Transcription start site (TSS): Where transcription begins, indicated numerically as position +1
Elongation: Mechanism of transcription, this is the reading of the template DNA and the formation of a polynucleotide RNA molecule
Termination: A mechanism of transcription, this is identification of the transcription stop site, dissociation of the transcriptional apparatus, and separation of the new RNA molecule from the DNA template
Consensus sequences: DNA sequences homologous in different genes of the same organism
Cis-acting elements: Consensus sequences located neat the region to be transcribed
Trans-acting factors: Proteins that influence gene expression by binding to cis-acting elements. One example is sigma factor in bacterial RNAP
Rho-independent termination: An enzyme traverses the entire gene until a termination sequence is encountered, self-complementary GC rich sequences form a hairpin which causes RNAP to stall adjacent to the poly U track, which as relatively weak interactions causing the RNAP to dissociate and release the transcript
Rho dependent termination: Termination in bacteria that relies on the rho dependent factor, which recognizes a binding site (rut) as soon as it is transcribed. Rho has helicase activity, and has a sequence to bind and cause the polymerase to stall and create a hairpin
Rho utilization site (rut): Where Rho binds to initiate the process of transcription ending
Rho: Binds to rut, will break hydrogen bonds and physically disassemble RNA polymerase
Hairpin secondary structure: Part of termination in bacteria, part of the newly formed transcript folds back in on itself forming this
Chromatin remodeling: The process of chromatin uncoiling to make DNA accessible to RNAP
RNA Polymerase I: Produces RNA, located in the nucleolus
RNA Polymerase II: Produces mRNA and snRNA, in the nucleoplasm, dependent of cis-acting elements and trans-acting transcription factors, binds to where the core-promoter is
RNA Polymerase III: Produces 5SrRNA and tRNA, located in the nucleoplasm
Proximal-promoter elements: Consist of enhancers and silencers, found upstream, within, or downstream of a gene, used in gene regulation and expression
Enhancer: Increases transcription levels
Silencer: Decreases transcription levels
General transcription factors (GTPs): Required by all RNAP II for transcription
Transcription activators and repressors: Influence efficiency or rate of RNAP II transcription initiation
TATA box: A core promoter element, binds TBP (TATA-binding protein) of transcription factor TFIID to determine the transcription start site
Pre-initiation complex: Formed by TBP binding to the TATA box and other transcription factors like TFDII binding allowing RNAP II to bind to it
DNA-RNA duplex: Part of RNA Pol II, bent at a right angle which positions the 3’ end of the RNA at the active site of the enzyme, allowing nucleotides to be added to the 3’ end
Abortive transcription: Transcription within RNA Pol II that is under 11 nucleotide pairs, due to it being initially unstable
Polyadenylation signal sequence: Signals termination in eukaryotes once detecting AAUAAA, the transcript is cleaves 10-35 base pairs further downstream
Rat1: An enzyme in eukaryotes for termination, works 5’→3’ with exonuclease activity. Attaches to a cleaved 5’ end of the RNA, moves down, and reaches the polymerase enzyme
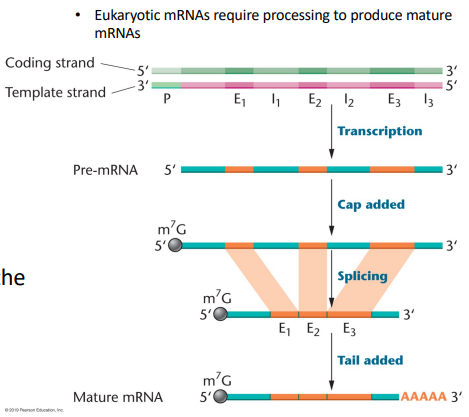
pre-mRNA: The transcript directly out of transcription, requires further processing. It is much longer and needs to be transported out of the nucleus and into the cytoplasm. Will get the addition of a 5’ cap (7-mg cap), addition of 3’ tail (poly-A tail), and have excision of introns and undergo slicing
Poly-A polymerase: Synthesizes a stretch of 250 adenylic acid residues at the 3’ end, attaches after the polyadenylation signal sequence (AAUAAA)
Poly A binding protein: Binds to the tail of pre-mRNA to protect it
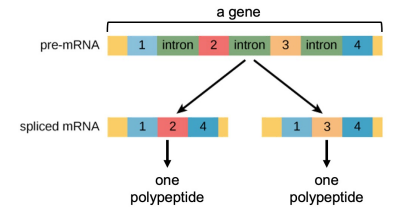
Introns: Regions of initial RNA transcription that aren’t expressed in amino acid sequences of the protein
Exons: Sequences retained and expressed, mostly present in only eukaryotes
Heteroduplexes: Introns present in DNA but not mRNA loop out
Untranslated regions (UTRs): RNA regions that will not be translated but regions that have other functions like binding to protein-making machinery
Splicing: The process that removes introns and joins exons together in mature mRNA
Heterogenous RNA (hnRNA): mRNA that contains all introns and exons, pre-splicing
Alternative RNA splicing: Allows for different segments to be treated as exons during RNA processing, the presence of introns allows a single gene to encode more than one kind of polypeptide
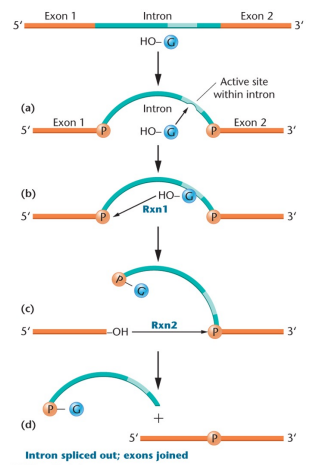
Self-splicing RNAs: The intron is the source of enzymatic activity and removes itself through self-excision, requires ribozymes
Ribozyme: An enzymatic RNA
Spliceosome: Where pre-mRNA introns are spliced out, uses reactions that involve formation of lariat structure, splice donor and acceptor sites, and branch point sequences
Prinbow box: A bacterial consensus sequence, example is TTGACA
In eukaryotes transcription occurs in the nucleus, in prokaryotes it is in the free cell. mRNA leaves the nuclease for translation
Transcription is initiated at RNA polymerase II promoters when TFIID and TBP transcription factors bind to the TATA box, followed by the binding of other general transcription factors, allowing RNAP II to bind to TFIID
The DNA double helix will enter RNA Pol II through a grove and unwind. Cleaving will destabilize RNAPII, opening up the clamp and releasing DNA and RNA