Campbell Unit 2: The Cell Cycle
Chapter 6: A Tour of the Cell
6.1: Biologists use microscopes and biochemistry to study cells
Light Microscope (LM): Visible light passes through a specimen and glass lenses, which refract the light in a way that magnifies the image of the specimen
Magnification: Image Size: Real Size ratio
Light microscopes magnify the image ~x1000
Resolution: Measure of clarity of the image, inversely related to wavelength of light a microscope uses for imaging
Organelles: Membrane enclosed compartments within cells
Electron Microscope (EM): Beam of electrons is shone through specimen or onto its surface
Scanning Electron Microscope (SEM): Electron beam scans surface of the sample which excites the surface elctrons. This is detected and translated into a 3D video
Useful for detailed study of topography of a specimen
Transmission Electron Microscope (TEM): Electron beam aimed through very thin section of the specimen which has been stained with atoms of heavy metals, which attach to certain cellular structures and thus enhance electron density in some parts more than others. Pattern of density is translated into an image
Used to study internal cell structure
Cell Fractionation: Take cells apart, separate major organelles and other subcellular structures from each other using a centrifuge (differential centrifugation)
Differential Centrifugation: Spins test tubes holding mixtures of disrupted cells at a series of increasing speeds
Cytology: Study of cell structure
Biochemistry: Study of chemical processes of cells
6.2: Eukaryotic cells have internal membranes that compartmentalize their functions
Prokaryotic Cells: Bacteria and archaea
DNA is in the nucleoid, which is not enclosed by a membrane
Eukaryotic Cells: Fungi, animals, and plants
Most DNA is in the nucleus, bounded by a double membrane
All cells…
Are bounded by a plasma/cell membrane
Have cytosol
Cytosol: Semifluid, jellylike substance where subcellular components are suspended
Have chromosomes with DNA
Have ribosomes
Ribosomes: Make proteins using gene instructions
Cytoplasm: Interior of any type of cell
In eukaryotic cells, this is only the region beyween the nucleus and plasma membrane
In some prokaryotic cells, there are regions surrounded by proteins in which specific reactions happen
Plasma Membrane: Selective barrier that allows passage of oxygen, nutrients, and wastes. A double layer of phospholipids and other lipids
6.3: The eukaryotic cell’s genetic instructions are house in the nucleus
Nucleus: Contains most genes in the eukaryotic cell
Nuclear Envelope: Encloses the nucleus and separates its components from the cytoplasm
Double membrane, each a lipid bilayer with associated proteins
Envelope has lots of pores. At the lip of each, the inner and outer membranes of the nuclear envelope are continuous
Pore complex (intricate protein structure) lines each pore and regulates entry and exit of proteins, RNAs, and large complexes of macromolecules
Nuclear Lamina: Netlike array of protein filaments that maintains shape of nucleus by mechanically supporting the nuclear envelope, lines nuclear side of the envelope except at the pores
Chromosomes: Discrete units that DNA is organized into, carry the genetic info
Chromatin: Complex of DNA and proteins making up chromosomes
Nucleolus: Mass of densely stained granules and fibers adjoining part of the chromatin to synthesize RNA
Nucleus directs protein synthesis by synthesizing mRNA → mRNA transported to cytoplasm via nuclear pores → translated by ribosomes into primary structure of a specific polypeptide once it reaches the cytoplasm
Ribosomes: Complexes made of ribosomal RNAs and proteins, carry out protein synthesis
Free Ribosomes: Ribosomes suspended in cytosol, which produce proteins that function within the cytosol
Bound Ribosomes: Outside the ER or nuclear envelope, produce proteins destined for insertion into membranes, packaging within organelles, or export from the cell (secretion)
6.4: The endomembrane system regulates protein traffic and performs metabolic functions
Endomembrane System: Synthesizes proteins, transports proteins and organelles into membranes or out of the cell, metabolism, movement of lipids, and detoxification of poisons
System includes nuclear envelope, endoplasmic reticulum, Golgi apparatus, lysosomes, vesicles and vacuoles, and plasma membrane
Vesicles: Sacs made of membrane, transfer membrane sacs
Endoplasmic Reticulum: Extensive network of membranes, accounts for 50%+ of the cell’s total membrane. Consists of cisternae, separates lumen from cytosol, continuous with the nuclear envelope
Cisternae: Membranous tubules and sacs
ER Lumen/Cisternal Space: Internal compartment of ER, cavity
Smooth ER: Outer surface has no ribosomes
Synthesizes lipids, metabolism, detoxifies drugs and poisons, stores calcium ions
Rough ER: Studded with ribosomes on the outer surface of the membrane
As a protein is built, it goes through a pore in the ER’s membrane, entering the lumen to take proper shape. Then the ER membrane keeps them separate, wrapped in membranes of vesicles, and are transported through transport vesicles
Glycoproteins: Most secretory proteins, proteins with carbohydrates covalently bonded to them
Rough ER grows in place by adding membrane proteins and phospholipids to its own membrane
Golgi Apparatus: Modifies and stores products, then sends them to other destinations. Kind of like a warehouse that receives, stores, ships, and even does some manufacturing
Group of associated flattened cisternae, looks like a stack of pita bread
Membranes of cisternae on opposite sides are different, cis and trans face
Cis face is near the ER, receives material from ER
Trans face gives rise to vesicles that pinch off and travel to other sites, transports material
Products are modified in transit between the two faces
Manufactures some macromolecules
ex. pectins and other noncellulose polysaccharides
Lysosome: Membranous sac of hydrolytic enzymes that are used to digest (hydrolyze) macromolecules, work best in acidic environments
Hydrolytic enzymes and lysosomal membrane are made by rough ER → Golgi for processing
Phagocytosis: Engulfing smaller organisms or food particles, how amoebas and other unicellular protists eat
Food vacuole then fuses with the lysosome and enzymes digest the food
Autophagy: Process where lysosomes use their hydrolytic enzymes to recycle their own organic material. Damaged organelle or small amount of cytosol is surrounded by a double membrane and a lysosome fuses with the outer membrane of it. Inner membrane and material dismantled by lysosomal enzymes, resulting compounds are released to be reused by cytosol
Vacuoles: Large vesicles derived from ER and golgi apparatus
Contractile Vacuoles: Pump excess water out of the cell
Central Vacuole: Large in mature plant cells, inside is cell sap (cell’s main repository of inorganic ions), plays major role in growth of plant cells which enlarge as it absorbs water
6.5: Mitochondria and chloroplasts change energy from one form to another
Mitochondria: Sites of cellular respiration
Cellular Respiration: Metabolic process that uses oxygen to drive generation of ATP by extracting energy from sugars, fats, and other fuels
Chloroplasts: Sites of photosynthesis in algae and plants
Endosymbiont Theory: Nonphotosynthetic prokaryotes engulfed a photosynthetic bacteria, as they developed together in symbiosis, it turned into mitochondria/chloroplasts
Double membranes in mitochondria, chloroplasts, and bacteria cells
Replication method is similar
Circular DNA
Mitochondria has a phospholipid bilayer with a unique collection of embedded proteins. Outer membrane is smooth, inner membrane is convoluted with cristae
Cristae: Infoldings
Intermembrane Space: Region between membranes
Mitochondrial Matrix: Second compartment enclosed by inner membrane, has many different enzymes, mitochondrial DNA, and enzymes
Thylakoids: Flattened, interconnected sacs in the chloroplast
Granum: Stack of thylakoids
Stroma: Fluid outside the thylakoid, has chloroplast DNA, ribosomes, and many enzymes
Plastids: Family of closely related plant organelles
Peroxisome: Spexialized metabolic compartment with a single membrane. Has enzymes that remove H atoms from substrates and transfer them to oxygen to produce hydrogen peroxide (H2O2)
Some use oxygen to break fatty acids down into smaller molecules
Ones in the liver detoxify alcohol and other harmful compounds
Has a enzyme that converts hydrogen peroxide to water
6.6: The cytoskeleton is a network of fibers that organizes structures and activities in the cell
Cytoskeleton: Network of fibers extending throughout the cytoplasm. 3 main types of fiber
Microtubules: Thickest cytoskeleton fiber, hollow rods, made from tubulin proteins (dimers, molecule made of 2 subunits)
Tubulin dimer consists of a and B tubulin
Plus end releases dimers at a higher rate
Microfilaments/Actin: Thin solid rods build from two intertwined strands of the globular protein actin (twisted double chain)
Present in all cells
Can form structural networks when certain proteins bind along the side of a filament to make branches
Immediate Filaments: Intermediate diameter (between microtubules and microfilaments), specialized for bearing tension, diverse class of cytoskeletal elements, permanent even after cells die
Centrosome: Region located near nucleus, where microtubules grow out of, the compression resisting girders of the cytoskeleton
Centrioles: Pair of them within the centrosome, each pair has 9 sets of triplet microtubules arranged in a ring
Flagella & cilia sometimes in eukaryotic cells, cellular extensions that contain microtubules
Flagella have just a few in a cell and are longer than cilia. Beats in undulating motion like a fishtail
Motile cilia are in large numbers on the cell surface and have alternating power and recovery strokes like oars. May also be like a signal recieving antenna for the cell (these are nonmotile and there is only 1 per cell)
Both have 9+2 pattern, 9 doublets of microtubules in a ring with two single ones in its center
Basal Body: Anchors cilia or flagellum microtubule assembly, structurally similar to a centriole
Dyenins: Large motor proteins attached along each outer microtubule doublet in flagella and motile cilia. Has two feet that walk along the microtubule of the adjacent doublet
Cortex: Outer cytoplasmic layer of a cell with a gel semisolid consistency
Myosin: Protein that makes up actin filaments and thicker filaments to cause contraction of muscle cells
Pseudopia: Cellular extensions that help a cell crawl along a surface
Cytoplasmic Streaming: Circular flow of cytoplasm within cells
6.7: Extracellular components and connections between cells help coordinate cellular activities
Cell Wall: Extracellular structure of plant cells, thicker than plasma membrane
Primary Cell Wall: Relatively thin and flexible wall secreted first by a young plant cell
Middle Lamella: Thin layer rich in sticky polysaccharides (pectins), between primary walls of adjacent cells, glues them together
Some cells strengthen cell walls by secreting hardening substances into the primary wall
Secondary Wall: Added between plasma membrane and primary wall with strong and durable matrix that affords the cell protection and support in cells that don’t secrete hardening substances into primary wall
Extracellular Matrix (ECM): Elaborate animal cell structure like a cell wall
Collagen: Forms strong fibers outside the cells, most abundant glycoprotein in the ECM, ~40% total protein in the human body
Embedded in a network of proteoglycans
Gibronectin: ECM glycoprotein, binds integrins to ECM
Integrins: Cell surface receptor proteins, span the membrane
Plasmodesmata: Channels that connect cells, perforate plant cell walls
Tight Junctions: Plasma membranes of neighboring cells are pressed very tightly against each other, bound by specific proteins
Desmosomes: Fasten cells together into strong sheets, attach muscle cells to each other
Gap Junctions: Provide cytoplasmic channels from one cell to an adjacent cell, create pores through which things may pass, necessary for communication between cells
6.8: A cell is greater than the sum of its parts
Many components work together in a functioning cell
Chapter 7: Membrane Structure and Function
7.1: Cellular membranes are fluid mosaics of lipids and proteins
Ampipathic: Has a hydrophillic and hydrophobic region
Fluid Mosaic Model: Membrane is a mosaic of protein molecules bobbing in a fluid bilayer of phospholipids
Integral Proteins: Penetrate hydrophobic interior of the lipid bilayer
Transmembrane Proteins: Span the membrane, majority of integral proteins
Others extend only partway into the interior
Peripheral Proteins: Not embedded in bilayer, loosely bound to surface
Glycolipids: Membrane carbohydrates covalently bonded to lipids
Glycoproteins: Membrane carbohydrates covalently bonded to proteins
7.2: Membrane structure results in selective permeability
Selective Permeability: Allows some substances to cross more easily than others
Transport Proteins: Help hydrophillic substances pass through the membrane
Aquaporins: Facilitates passage of water molecules through the membrane
7.3: Passive transport is diffusion of a substance across a membrane with no energy investment
Diffusion: Movement of particles from high to low concentration
Concentration Gradient: Region along which the density of a substance increases or decreases
Passive Transport: Transport that requires no energy
Osmosis: Diffusion of free water across a selectively permeable membrane
Tonicity: Ability of a surrounding solution to cause a cell to gain or lose water
Hypotonic: Solution has less solutes than in another
Water is hypotonic to everything.
Water enters cells.
Animal cell is Lysed 😢
Plant cell is Turgid 🙂
Isotonic: Equal number of solutes in both solutions
Animal cell is Normal 🙂
Plant cell is Flaccid 😐
Hypertonic: Solution has more solutes than another
Water leaves cells.
Animal cell is Crenate 😢
Plant cell is Plasmolysed 😢
Osmoregulation: Control of solute concentrations and water balance in organisms without rigid cell walls
Facilitated Diffusion: Polar molecules and ions diffuse passively with the help of transport proteins
Ion Channels: Channel proteins that transport ions
Gated channels: Open or close in response to a stimulus
7.4: Active transport uses energy to move solutes against their gradients
Active Transport: Using ATP to transport a solute across a membrane
Enables internal concentrations to differ from external concentrations
ex. Sodium potassium pump
Membrane Potential: Voltage across a membrane
Cytoplasmic side of a membrane is negative in charge relative to the extracellular side
Ranges from -50 to -200 mV
Electrochemical Gradient: Combination of chemical force and an electrical force acing on an ion
Electrogenic Pump: Transport protein that generates voltage across a membrane
Proton Pump: Actively transports protons out of the cell
Cotransport: Using proton gradient to power something else
7.5: Bulk transport across the plasma membrane occurs by exocytosis
Exocytosis: Secreting certain molecules by fusion of vesicles with the plasma membrane
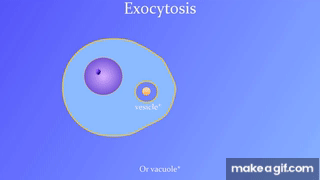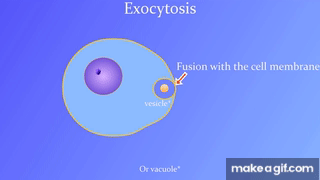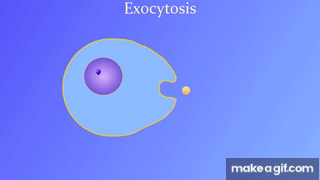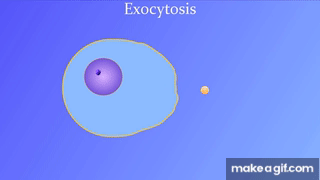
Endocytosis: Cell takes in molecules by forming new vesicles by pinching off part of the membrane.
Phagocytosis: Cell engulfs a particle by extending pseudopia around it and packaging it in a food vacuole
Pinocytosis: Cell continually “gulps” droplets of extracellular fluid into tiny vesicles
Receptor Mediated Endocutosis: Specialized type of pinocytosis, enables cell to aquire large quantitites of specific substances

Chapter 8: An Introduction to Metabolism
8.1: An organism’s metabolism transforms matter and energy
Metabolism: Totality of an organism’s chemical reactions
Metabolic Pathway: Specific molecule is altered in a series of defined steps, each catalyzed by a specific enzyme, resulting in a certain product
Catabolic Pathways: Breakdown pathways, release energy
Anabolic Pathways: Consume energy to build complex molecules from simpler ones
Kinetic Energy: Energy associated with relative motion of objects
Thermal Energy: Kinetic energy associated with random movement of atoms or molecules
Heat: Thermal energy in transfer from one object to another
Potential Energy: Energy that matter posesses because of its location or structure
Chemical Energy: Potential energy available for release in a chemical reation
Spontaneous Process: Process that can happen without input of energy, leads to an increease in entropy by itself
Thermodynamics: Study of energy transformatios that occur in a collection of matter
First Law of Thermodynamics/Principle of Conservation of Energy: The energy of the universe is constant—Energy can be transferred and tranformed but not created or destroyed
Second Law of Thermodynamics: Every energy tranfer or transformation increases the entropy of the universe
Entropy: Measure of molecular disorder/randomness
8.2: The free-energy change of a reaction tells us whether or not the reaction occurs spontaneously
Free Energy: Portion of energy that can perform work when temperature and pressure are uniform
ΔG = ΔH - TΔS
ΔG is change in free energy
ΔG = Gfinal state - Ginitial state
ΔH is change in system’s enthalpy (total energy)
ΔS is change in system’s entropy
T is absolute temperature in Kelvins
Exergonic Reaction: Net release of free energy, negative ΔG
Endergonic Reaction: Absorbs free energy from its surroundings, positive ΔG
8.3: ATP powers cellular work by coupling exergonic reactions to endergonic reactions
Three main kinds of work that a cell does
Chemical Work: The pushing of endergonic reactions that would not occur spontaneously
ex. Synthesis of polymers from monomers
Transport Work: Pumping of substances across membranes against direction of spontaneous movement
Mechanical Work
ex. Beating of cilia contraction of muscle cells, movement of chromosomes during cellular reproduction
Energy Coupling: The use of an exergonic process to drive an endergonic one, how cells manage energy resources to do work
ATP (Adenosine Triphosphate): Sugar ribose + nitrogenous base adenine + chain of 3 phosphate groups (like a compressed spring)
Terminal phosphate bond can be broken by hydrolysis, when it is, splits into inorganic phosphate molecule (HOPO32-or Pi) and ADP
ATP + H2O —> ADP + Pi
ΔG = -7.3 kcal/mol (-30.5 kJ/mol)
Phosphorylated Intermediate: Recipient molecule of phosphate group from ATP, which is made less stable
ATP is renewable, and can be regenerated by the addition of phosphate to ADP
Free energy required to phosphorylate ATP comes from exergonic catabolism in the cell
Chemical potential energy stored in ATP drives most cellular work
It uses energy from catabolism to phosphorylate ADP and generate ATP
First one since second one involves the synthesis of ATP, which requires energy, so it is endergonic, which means free energy is positive
active
8.4: Enzymes speed up metabolic reactions by lowering energy barriers
Enzyme: Macromolecule that acts as a catalyst
Catalyst: Chemical agent that speeds up a reaction without being consumed by it
Activation Energy: Energy required to contort reactant molecules so bonds can break—amount of energy needed to push the reactants to the top of a barrier so the downhill part can begin
Lowered by enzymes
Substrate: Reactant an enzyme acts on
Enzyme Substrate Complex: Enzyme binded with substrate
Active Site: Restricted region of the enzyme that actually binds to the substrate
Induced Fit: Tightening of binding after initial contact
Cofactors: Nonprotein helpers for catalytic activitym either bound tightly to the enzyme or bound loosely and reversible along with the substrate
Coenzyme: Organic cofactor
Competitive Inhibitors: Reversible inhibitors, resemble normal substrate molecule, compete for admission into the active site
Noncompetitive Inhibitors: Bind to another part of the enzyme that changes the shape of the active site
8.5: Regulation of enzyme activty helps control metabolism
Allosteric Regulation: Used to describe any case where protein’s function at one site is affected by binding of a regulatory molecule at another
Cooperativity: Substrate molecule binds to one active site in a multisubunit enzyme, triggers a shape change, increasing catalytic activity at other active sites, amplifies response of enzymes to substrates
One substrate primes an enzyme to act on other substrates more readily
Feedback Inhibition: Metabolic pathway halted by inhibitory binding of its end product to an enzyme that acts early in the pathway
Chapter 9: Cellular Respiration and Fermentation
9.1: Catabolic pathways yield energy by ocidizing organic fuels
Fermentation: Partial degredation of sugars or other organic fuels without ocygen
Anabolic Respiration: Oxygen consumed as a reactant along with organic fuel
Anaerobic Respiration: Oxygen is not the final oxidizing substance, used by some prokaryotes
Cellular Respiration: Used to refer to aerobic process
C6H12O6 + 6O2 —> 6CO2 + 6H2O + Energy (ATP + heat)
Glucose breakdown is exergonic, free energy change of ΔG = -686 kcal/mol
Redox Reactions: Oxidation-Reduction reactions, electron transfers
Oxidation: Loss of Electrons
Oxidizing Agent: Electron donor
Reduction: Addition of Electrons
Reducing Agent: Electron acceptor
LEO the lion says GER!
Loss of Electrons Oxidation, Gain of Electrons Reduction
Redox reactions can also change the degree of electron sharing in covalent bonds
Glucose is broken down in a series of steps catalyzed by enzymes
Electrons stripped from glucose at key steps
Each electron travels with a proton as a hydrogen atom
NAD is used as an electron carrier because it can easily cycle through NAD+ and NADH
Electron Transport Chain: Mostly proteins built into the inner membrane of the mitochondria (this is in eukaryotes, in prokaryotes, it’s the plasma membrane), a ladder with higher and lower energy
Three stages of cellular respiration: Glycolysis, Pyruvate Oxidation + Citric Acid Cycle, and Oxidative Phosphorylation
Glycolysis: Breaks glucose into two molecules of pyruvate, which enters the mitochondrion and is oxidized into acetyle CoA, which goes into the citric acid cycle, occurs in the cytosol
Citric Acid Cycle: Breakdown of glucose into carbon dioxide (in prokaryotes, this is in the cytosol)
Oxidative Phosphorylation: Electrons combined with O2 and H+, forming water. Energy released by each step helps turn ADP into ATP. Powdered by redox reactions of the ETC
Accounts for almost 90% of the ATP generated by respiration
Substrate Level Phosphorylation: Enzyme transfers phosphate group from substrate to ADP
Aerobic respiration generates much more ATP since oxidative phosphorylation accounts for almost 90% of ATP produced by the cell
not sure
9.2: Glycolysis harvests chemical energy by oxidizing glucose to pyruvate
2 phases, energy investment and energy payoff
Net energy yield is 2 ATP and 2 NADH per glucose
Means “sugar splitting”, since glucose is split into 2 three carbon sugars, whihc are oxidized and rearranged to form pyruvate
9.3: After pyruvate is oxidized, the citric acid cycle completes the energy-yielding oxidation of organic molecules
Acetyl Coenzyme (acetyl CoA): When entering mitochondria via active transport, pyruvate first converted into this
Pyruvate’s carboxyl group is fully oxidized and given off as a molecule of CO2
Remaining two carbon fragment is oxidized, electrons transferred to NAD+ (now NADH)
Coenzyme A (CoA) attached to two carbon intermediate, forming acetyl CoA
Citric acid cycle has 8 steps, each catalyzed by a specific enzyme
Acetyl CoA adds its acetyl group to oxaloacetate to make citrate
Citrate converted to isocitrate, its isomer, by removing one water molecule and adding another
Isocitrate is oxidized, reducing NAD+ to NADH. This compound loses a CO2
Another CO2 molecule is lost, and the compound is oxidized, reducing NAD+ to NADH
CoA is displaced by a phosphate group, which is transferred to GDP, forming GTP (similar to ATP, can also be used to generate it)
Two hydrogens transferred to FAD to form FADH2, oxidizing the old compound
Addition of water molecule rearranges bonds
Substrate is oxidized, reducing NAD+ to NADH
Oxaloacetate is made up of different carbon atoms each time around
3 NAD+ reduced each time
When NAD+ is reduced, it is exothermic
Citric Acid Cycle
Exothermic reations used to power endothermic ones
9.4: During oxidative phosphorylation, chemiosmosis couples electron transport to ATP synthesis
In the inner mitochondrial membrane in eukaryotic cells (plasma membrane of prokaryote)
Electrons acquired from glucose by NAD+ are transferred to first molecule of ETC in complex I
This is a flavoprotein, which returns to its oxidized form by passing electrons to an iron sulfur protein (Fe-S in complex I)
Then passed to ubiquinone, a small hyrophobic molecule that;s not a protein
Remaining carriers are cytochromes
Cytochrome: Protein that accepts and donates electrons
Cyt3 passes electrons to O2 which is very electronegative, then picks up protons from the aqueous solution, forming water
FADH2 also donates electrons to complex II, but there is 1/3 the amount of ATP produced
Chemiosmosis: Process where energy stored in the hydrogen ion gradient across a membrane is used to drive cellular work (such as ATP synthesis)
Proton Motive Force: H+ gradient
Most energy flows from glucose —> NADH —> ETC —> proton motive force —> ATP
Around 30 to 32 ATP per glucose (2 glycolysis (substrate level phosphorylation), 2 ATP (substrate level phosphorylation), 26 or 28 (oxidative phosphorylation)
Ratio of NADH to ATP is not whole (phosphorylation and redox reactions not directly coupled to each other), around 4+ H+ reenter mitochondria via ATP synthase
Single NADH has enough proton motive force for 2.5 ATP
FADH2 only enough for 1.5 ATP
Chapter 10: Photosynthesis
10.1: Photosynthesis feeds the biosphere
Photosynthesis: Conversion turning energy of sun and light into chemical molecules
Acquires organic compounds through autotrophic nutrition or heterotrophic nutrition
Autotrophs: “self feeders”, sustain selves without eating anything from other living beings
The producers, use CO2 and inorganic raw materials
Heterotrophs: Live on compounds from other organisms
Decomposers: Eat remains of other organisms and feces
They consume autotrophs to sustain themselves
The algae can then produce more oxygen from the excess carbon dioxide
10.2: Photosynthesis converts light energy to the chemical energy of food
Endosymbiont Theory: Chloroplast was from the cyanobacteria being engulfed by a eukaryotic cell, and they began to undergo symbiosis, same as mitochondria
Chloroplast: Eukaryotic organelle that absirbs sunlight to drive synthesis of organic compounds
Found mainly in cells of mesophyll
Mesophyll: Tissue in interior of leaf, typically has 30-40 chloroplasts (each 2-4 µm by 4-7 µm)
Stomata: Microscopic pores where CO2 enters and O2 exits
Stroma: Dense fluid, surrounded by two membranes
Thylakoids: In stroma, segregates it from the thylakoid space
Grana: Columns of thylakoids
Chlorophyll: Green pigment that gives leaves their color, in the membranes of thylakoids
6CO2 + 12H2O + Light Energy —> C6H12O2 + 6H2O + O2
Two parts of photosynthesis
Light Reactions (photo): Convert solar energy to chemical energy
Water is split, providing a source of electrons and protons, and giving O as a byproduct
Light absorbed by chlorophyll drives transfer of electrons and hydrogen ions from water to NADP+
Solar energy used to reduce NADP+ to NADPH by adding a pair of electrons and H+
Uses chemiosmosis to power addition of phosphate group to ADP
Photophosphorylation: Add phosphate group to ADP
Light energy converted to chemical energy as NADPH and ATP
NADPH as “reducing power” that can be passed along to electron acceptors
Calvin Cycle: Turns carbon dioxide into sugar
Carbon Fixation: Incorporating CO2 from air into organic molecules already present
Reduces fixed carbon into carbohydrate by reducing it (using NADPH and ATP)
In the stroma
Idk lol
What? Too many big words
The Calvin cycle provides NADP+ and ADP, as well as produces glucose for energy.
10.3: The light reactions convert solar energy to chemical energy of ATP and NADPH
Light is a form of electromagnetic energy
Wavelength: Distance between crests
Electromagnetic Spectrum: Range of radiation
Visible light: (purple) 380 nm — 740 nm (red) wavelength, colors
Photons: Particles of light
Pigments: Substances that absorb visible light
Spectrophotometer: Measures ability to absorb different wavelengths of light
Chlorophyll a: Light capturing pigment that participates directly in light reactions
Chlorophyll b: Accessory pigment
Carotenoid: Hydrocarbons, may broaden spectrum of colors that drive photosynthesis, photoprotection
Photoprotection: To absorb and dissapate excessive light energy that would otherwise damage chlorophyll or form oxidadive molecules that are dangerous to the cell
Action Spectrum: Profiles relative effectiveness of different wavelengths of radiation
Absorption of a photon boosts an electron to a ring further than to the nucleus, meaning higher potential energy, and this is their excited state
Only photons absorbed are those with energy equal to energy diff between ground and excited state of electron, so only specific wavelengths
Can only stayfor a billionth of a second, releasing excess energy as photon and fluorenscence (afterglow) happens
Photosystem: Composed of reaction center complex surrounded by light complexes
Reaction Center Complex: Organized association of proteins with a special pair of chlorophyll a molecules and a primary electron acceptor, purpose to convert light to energy
Primary Electron Accepter: Molecule capable of accepting electrons and being reduced
Light Harvesting Complex: Pigment molecules bound to proteins, traps light to transfer to the reaction center
First step of light reaction, solar powered transfer of electron from the reaction center chlorophyll a pair to the primary electron acceptor
When chlorophyll electron excited, primary electron acceptor captures it in a redox reaction
Two types of photosystems in the thylakoid membrane
Photosystem II (PSII): Reaction center chlorophyll a called P680, best at absorbing light with a wavelength of 680 nm (red).
P680 is the strongest biological oxidizing agent known, and its electron “hole” mmust be filled
Photosystem I (PSI): Reaction center chorophyll a called P700, best at absorbing light with a wavelength of 700 nm (far red)
Linear Electron Flow: Flow of electrons through photosystems and other molecular components from the thylakoid membrane
A photon of light strikes one of the pigment molecules, exciting one of its electrons.
As it falls back down, an electron in a nearby pigment is excited, and this keeps going until it reaches the P680.
An electron in the pair of cholophyll a is excited
This electron is transferred from the excited P680 to the primary electron acceptor, and P680 is now P680+
Enzyme splits water molecule into two electrons, two hydrogen ions, and an oxygen atom
Electrons are supplied one by one to the P680+ pair, each electron replacing one transferred to the primary electron acceptor
H+ released into thylakoid space
Oxygen atom combines with another oxygen atom split from water, forming O2
Photoexcited electrons go from PSII to PSI using an electron transport chain (made of electron carrier plastoquinone and protein plastocyanin). Each component carries out redox reactions as electrons flow down
This releases free energy used to pump H+ into thylakoid space and make a proton gradient
Potential energy in proteon gradient is used to make ATP through chemiosmosis
Light energy captured through light harvesting pigments to the PSI, exciting a P700 electron
Photoexcited electron transferred to PSI’s primary electron acceptor, making it P700+, with a hole, so it accepts an electron from the bottom of the electron transport chain from PSII
Photoexcited eectrons go to second electron transport chain (through protein ferredoxin)
NADP+ reductae catalyzes fransfer of electrons to NADP+, which needs two electrons to be reduced into NADPH and also removes an H+ from the stroma
Cyclic Electron Flow: Uses photosystem I but not photosystem II, alternate path. Electrons go from ferredoxin to cytochrome complex, them using plastocyanin to P700. Generates ATP but no production of NADPH or release of oxygen
Green, since all pigments absorb green the worst, and reflect/transmit it instead, hence the green appearance of the plant
H20, NADP+
No ATP is created
10.4: The Calvin Cycle uses the chemical energy of ATP and NADPH to reduce CO2 to sugar
In the stroma
Similar to citric acid cycle, since the starting material is regenerated as some molecules enter and exit the cycle
G3P (Glyceraldehyde 3 phosphate): Carbohydrate produced directly from Calvin cycle, cycle must take place 3 times to synthesize 1
Carbon Fixation: Phase 1 of the Calvin cycle. Rubisco attaches CO2 with ribulose biphosphate (RuBP), a five carbon sugar. It forms an unstable 6 carbon compound before splitting into 2 molecules of 3-Phosphoglycerate
Rubisco: Enzyme that joins RuBP and CO2, the most abundant protein in chloroplasts & on earth
Phase 2 is reduction. Each molecule gets a phosphate group from ATP (which becomes ADP) and a pair of electrons from NADPH (which becomes NADP+).
In the last phase, regeneration, 1 G3P exits the cycle, while the other 5 regenerate into RuBP, which consumes 3 ATP in the process.
10.5: Alternative mechanisms of carbon fixation have evolved in hot, arid climates
C3 Plants: CO2 combined with RuBP using rubisco
Rubisco can also bind O2 instead of CO2, producing CO2 and using ATP instead of making it
Close stomatas on hot, dry days
Photorespiration: Consumes O2 (photo), produces CO2 (respiration) using ATP and not producing sugar
Then uses the generated CO2 for regular carbon fixation
C4 Plants: Alternate form of carbon fixation before Calvin cycle that produces a four carbon compound as first product
Evolved independently at least 45 seperate times. More efficient than C3 since it uses less water and resources.
When weather is hot and dry, partially closes stomata
Photosynthesis starts in mesophyll cells but is completed in bundle sheat cells
Bundle Sheath Cells: Cells arranged into tightly packed sheaths around the veins of the leaf
PEP Carboxylase: Higher affinity for CO2 than rubisco, no affinity for O2
PEP Carboxylase adds CO2 to PEP, producing ocaloacetate, even when lower CO2 concentration and relatively higher O2 concentration
Four carbon products exported to bundle sheath cells through plasmodesmata
Within bundle sheath cells, an enzyme releases CO2 and it is refixed by rubisco and the Calvin cycle
This also regenerates pyruvate in the same wau, which is transported to the mesophyll cells (ATP converts pyruvate into PEP)
Crassulacean acid metabolism (CAM): Carbon fixation mode, during night plants use CO2 in orgaic acids (stored in mesophyll cells). During day stomatas closed, and CO2 is released for use in Calvin Cycle
Turned into ATP through cellular respiration
Chapter 11: Cell Communication
11.1: External signals are converted to responses within the cell
Earl W Sutherland investigated how animal hormone epinephrine leads to glycogen breakage
Cellular communication has three stages in the signal transduction pathway
Signal Reception: Cell’s detection of a signaling molecule coming from outside the cell, when the signaling molecule binds to a receptor protein at the cell’s surface
Signal Transduction: Binding of signaling molecule changes the receptor protein and initiates transduction. Converts signal to a form that brings a specific cellular response
Cellular Response: Transduced signal triggers a cellular response
11.2: Signal reception — A signaling molecule binds to areceptor, causing it to change shape
Ligand: Molecule that specifically binds to another
Signaling molecule has a complementary shape to a site on the receptor and binds there, acting as a ligand
G Protein Coupled Receptor: Cell surface transmembrane receptor that works with the help of a G Protein
G Protein: Protein that binds energy rich GTP
Receptor Tyrosine Kinases (RTKs): Plasma membrane receptor with enzymatic activity, a protein kinase (enzyme that catalyzes transfer of phosphate groups from ATP to another protein)
Ligand Gated Ion Channel: Membrane channel receptor with a region that acts as a “gate” and opens or closes when the receptor changes shape
11.3: Signal transduction — Cascades of molecular interactions transmit signals from receptors to relay molecules in the cell
Protein Kinase: Enzyme that tranfers phosphate groups from ATP to a protein
Phosphorylation Cascade: Series of different proteins in a pathway are phosphorylated and add a phosphate group to the next one in line
Protein Phosphatases: Enzymes that can rapidly dephosphorylate (remove phosphate groups from) proteins
Second Messengers: Small nonprotein water soluble molecules or ions which can easily spread through regions of the cell by fiddusion and particpate in participate in pathways initiated by G protein coupled receptors and receptor tyrosine kinases
Cyclic AMP (cAMP): Small molecule produced from ATP
Adenylyl Cyclase: Converts ATP to cAMP in response to an extracellular signal
Inositol Triphosphate (IP) and diacylglycerol (DAG) also lead to calcium release
11.4: Cellular response — Cell signaling leads to regulation of transcription or cytoplasmic activities
Some pathways lead to a nuclear response—specific genes are turned on or off by activated transcription factors
In others the response involves cytoplasmic regulation
Cellular repsonses are regulated at many steps
Each protein in a signaling pathway amplifies the signal by activating mutliple copies of the next component
For long pathways the total amplification may be over a millionfold
Scaffolding Proteins: Large relay proteins to which many other relay proteins are simultaneously attached to, permanently hold together networks of signaling proteins at synapses. Increase signaling efficiency
Pathway branching further helps the cell to coordinate signals and responses
11.5: Aptosis requires integration of multiple cell signaling pathways
Aptosis: Controlled cell suicide, cellular agents chop up DNA and fragment organelles and other cytoplasmic components
Chapter 12: The Cell Cycle
12.1: Most cell division results in genetically identical daughter cells
Cell Division: Reproduction of cells, distinguishes living things from nonliving
Helps with asexual reproduction, growth/development, and tissue renewal
Genome: Cell’s DNA or genetic information
Human cell usually has about 2m of DNA, 250,000x the cell’s diameter
Chromosomes: DNA are packaged into these structures
Chromatin: The DNA and proteins that is the building material of chromosomes
Somatic Cells: All cells that aren’t the reproductive cells, contain 46 chromosomes (2 sets of 23)
Reproductive Cells: Have half as many chromosomes as somatic cells (1 set of 23)
Each duplicated chromosome has two sister chromatids with identical DNA molecules, attached along their lengths by cohesins
This is called sister chromatid cohesion
Centromere: Where the chromatid is most closely attached to the other one, the “waist”
When the two sister chromatids seperate, they are now individual chromosomes
Mitosis: Division of genetic material in the nucleus
Cytokinesis: Division of the cytoplasm, right after mitosis
Meiosis: Produce gametes(male sperm cell or female egg cell) through it, modified version of mitosis
In step 1, there is one. In step 2, there is one. In step 3, there are two.
39, 39, 39
12.2: The mitotic phase alternates with interphase in the cell cycle
Cell Cycle: Life of a cell from when it’s formed until division
Mitotic (M) Phase: Mitosis + Cytokinesis, usually shortest part of cell cycle
5 stages: prophase, prometaphase, metaphase, anaphase, and telophase
Prophase — pro, first, where nucleus is still there, chromosomes condensing
Metaphase — middle, where the chromosomes line up, nucleus gone
Anaphase — away, chromatids separated and moving away from middle (towards poles/centrioles)
Telophase — two, nuclei begin to form and chromosomes at complete opposite ends
Interphase: ~90% of cell cycle, contains G1 (1st gap) phase, S (synthesis) phase, and G2 (2nd gap) phase
Cell grows (G1), keeps growing and copies it’s chromosomes (S), continues to grow as it finishes preparing for cell division (G2), and divides (M).
Mitotic Spindle: Forms in cytoplasm during prophase. Other microtubules partially disassemble to form it. They polymerize (elongate) by adding more subunits of tubulin, a protein, and shorten by losing them
Includes centrosomes, spindle microtubules, and asters
Centrosome: Where spindle assembly starts, also organizes microtubules and provides structure
Pair of centrioles are at the center of the centrosome
During interphase in animal cells, it duplicates and forms two. They move apart during prophase and prometaphase (the two p’s) as spindle microtubules grow from them
By the end of prometaphase, they’re at opposite ends of the cell
Aster: Radial array of short microtubules, comes from each centrosome
Kinetochore: Structure of proteins, what the spindle attaches to. Each sister chromatid has one
Metaphase Plate: Imaginary plate, where centromeres of duplicated chromosomes are at metaphase (middle)
In animal cells, cytokinesis is through cleavage
Cleavage Furrow: Shallow groove in cell surface near old metaphase plate
In plant cells, vesicles move along microtubules to middle of cell, where they coalesce (combine), making a cell plate
Binary Fission: Reproduction where prokaryotic cell grows to double its size, then divides into two cells
Origin of Replication: Where DNA of chromosome replicates, producing 2 origins
6
In animals, there is a cleavage, in plants, there is only a cell plate
Prophase, prometaphase, metaphase
idk
It connects to the centromeres, pulling the centrioles apart.
idk
12.3: The eukaryotic cell cycle is regulated by a molecular control system
Hypothetical evolution of mitosis is that it evolved from binary fission
This is because protists exhibit cell division between fission and meiosis
Cell Cycle Control System: Molecules in cell that trigger and coordinate key events
Checkpoint: Cell’s control point to see if everything is working
Cyclin: Kinases (Enzymes that catalyze transfer of phosphate group) must attach to them to be active, a family of proteins
Cyclin Dependent Kinases (Cdks): The kinases attached to cyclins
Maturation/M-Phase Promoting Faction (MPF): First discovered cyclin-Cdk complex, triggers passage into M Phase
In anaphase, switches itself off by destroying its own cyclin
3 important checkpoints at G1, G2, and M
G1checkpoint is usually considered most important, since it checks if the cell is fit for division. If doesn’t pass, it goes to G0 phase, kind of like a limbo.
In G, the cell is not actively dividing and does its normal functions. It can be “called back” into G.
G2 checkpoint makes sure all chromosomes have been replicated and replicated DNA is not damaged
If reparable, mitosis paused and it is fixed
If not, apoptosis (programmed cell suicide)
M checkpoint checks if sister chromatids are correctly attached to spindle, and is near the end of metaphase
When all chromosomes properly attached to spindle fibers, it proceeds into anaphase
Growth Factor: Protein released by some cells to tell other cells to divide
Density Dependent Inhibition: Crowded cells stop dividing
Anchorage Dependence: To divide, it must be attached to something
In cancer cells, they don’t stop dividing when growth factors are depleted
Benign Tumor: Abnormal cells remain at original site
Malignant Tumor: Spread to new tissues and impair functions of other tissues
Metastasis: Spread of cancer cells to distant locations from original site
It hasn’t passed through the S stage, synthesis
CDK and cyclin combine into MPF
idk