lecture 1: applications of cryo-electron microscopy to understand complex structures
Section A - revision of 3D cryo-EM and 3D projections
there are three main uses of cryo-em:
to gain high resolution native state 3D structure of proteins
to get 3D structure of unique cellular structures using electron tomography - tools for biochem year 2
correlating electron and light microscopy (CLEM)
gaining high resolution native 3D structure is focused on
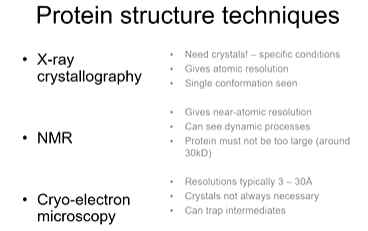
In special cases, 1 angstrom resolution in cryo-em can be achieved. This means it is more likely to get a structure from a challenging case. As samples are frozen on a quick time frame, intermediates can be trapped - polymerisation process can be viewed.
To get protein structure from single protein molecules, single particle analysis is used. Here, pictures of individual molecules are taken and are collected digitally and analysed. This means there is no need for crystals in order to determine protein structure
two methods of staining can be done in cryo-em:
negative stain - continuous carbon
cryo (holey film)
negative stain
This is not cryo-em but is used preamble to cryo em. The sample is applied to the electron microscopy grid and dried on the grid sample holder. A heavy metal stain is used - often urinal acetate - which allows a high contrast image to be achieved. This method is quick and easy
cryo
the support film has holes in it and the sample is put onto it as a liquid and the sample is rapidly blotted and frozen. This means that samples are suspended in the ice. Pictures can then be taken. The sample must be kept at -150 degrees to prevent ice crystals from forming.
the frozen sample is taken and put into an electron microscope where individual molecules are viewed. Contrast is very important and highly contrasted images are easier to obtain however, it is not used for structural biology
Ice is a good fixative and so chemicals and drying is not needed and the ice layer is vitreous and glass like. This also keeps the protein hydrated. When pictures are taken, the images are composed of phase contrast. However, the biological samples are destroyed by the electron beam through radiation damage.
In cryo-em, the source of electrons passes through the sample (transmission) and are weakly scattered as they are going through a sample that can’t scatter strongly as there are no heavy metals and the sample is thin. This means that when they coincide at the image, there is a mathematical relationship which is a close approximation of a 2D projection of the original sample.
Section B - what are class averages and how do we use them
The 2D projection is related to class averages which contains some 3D information of the sample. To get a complete image, the sample must be rotated fully and flattened. Enough of the 2D projections means that the structure can be rebuilt to get the 3D structure,
This is hard however, as low contrast images are often obtained. Class averages are used to get a high resolution structure,
Images that are the same are found and aligned and averaged. This means that you intensify the contrast as it has been averaged out. After this, the protein structure must be put back together. The angle the protein is at when it was flattened must be aligned so that they match and this is the aligning process.
For 3D cryo-em, many images of protein molecules are collected at different orientations are collected. The ones that are similar to each other are classified together however they might look similar but won’t be aligned rotationally. This means they must be aligned and this is done in the computer. This is rotational alignment so they can be averaged together. Sometimes, particles are assigned to the wrong class and this can happen is the views are similar. Other times, one class is too small and too many in one orientation (preferred orientation). This can be a problem as one class can be over represtated and one not as well represented and this will mess the class average and therefore the 3D structure.
A single image from each class is obtained with a greater contrast. If one part of the protein is mobile it can be hard to identify. As well as the protein sometimes being narrow and this won’t show up in the image in the ice.
Section C - making a 3D structure from 2d projections
Sometimes, images can be different sizes and so may not be included.
Some methods to determine orientations are:
angular reconstitution
uses the theory of common lines to determine relative orientations if single particles
projection matching
take a 3D model as a template to determine orientations by cross creation
analyse FT of ordered sample (tools for biochem)
projection matching
The structure that is thought to be similar is taken and 2D projections are made in the computer. This means that orientations are known. Cross correlation between the two images is done and the ones that are matched the best can have angels assigned to them. These angles then mean that the images can be aligned around the structure.
Now the 3D structure can be determined. The images can be unflattened in the computer and work out how they line up is already known. This means that a reconstruction can be done. Back projection is done where the 2D image is taken and the image data is built on top of 2D lines so make a 3D tower. At the right angle with every image, this makes a 3D structure. As the lines intersect is where the structure is.
This how the NOMPC ion channel was obtained. This is a pressure sensor in the protein membrane. This had to be purified first. Amphipolis can be used which has both hydrophilic and phobic properties to keep the protein in its native state. Nano discs can also be used which are lipids that peptides that stabilize the protein by keeping hydrophobic areas soluble. Initially, amphipolis seemed to have higher resolution but when negative stain was used and individuals particles were analysed, non discs gave better class averages. Experimenting with different conditions improved resolution when samples were obtained from both however, nanodiscs gave the best resolution to a much higher degree as nanoparticles likely reduced mobility the best as it is assumed that two images have the complete same molecular structure. This was enough to make a molecular structure.