8-Translation
Translation Overview
Location: Translation occurs in the cytosol.
Mechanism: mRNA base pair sequences are translated into amino acid sequences.
Codon Reading: Nucleotides are read in groups of three, forming codons.
There are 64 codons (4 different bases x 3 bases per code = 4³).
Each codon specifies a start, stop, or specific amino acid.
Genetic Code: The relationship between a codon and its corresponding translation is known as genetic code.
tRNA Molecule
Function: tRNA acts as the link between codons and their corresponding amino acids.
Structure: tRNA is approximately 80 nucleotides in size with an anticodon region that binds to the mRNA codon.
tRNA Synthesis and Modifications
Synthesis: tRNA is synthesized by RNA Polymerase III as large transcripts which are subsequently trimmed. —> trimmed at ends and in middle
Some bases undergo chemical modifications to yield unique non-traditional bases (e.g., Ψ, Im, Gm, and others).
Conformation: These modifications define distinct final shapes of the tRNA molecules.
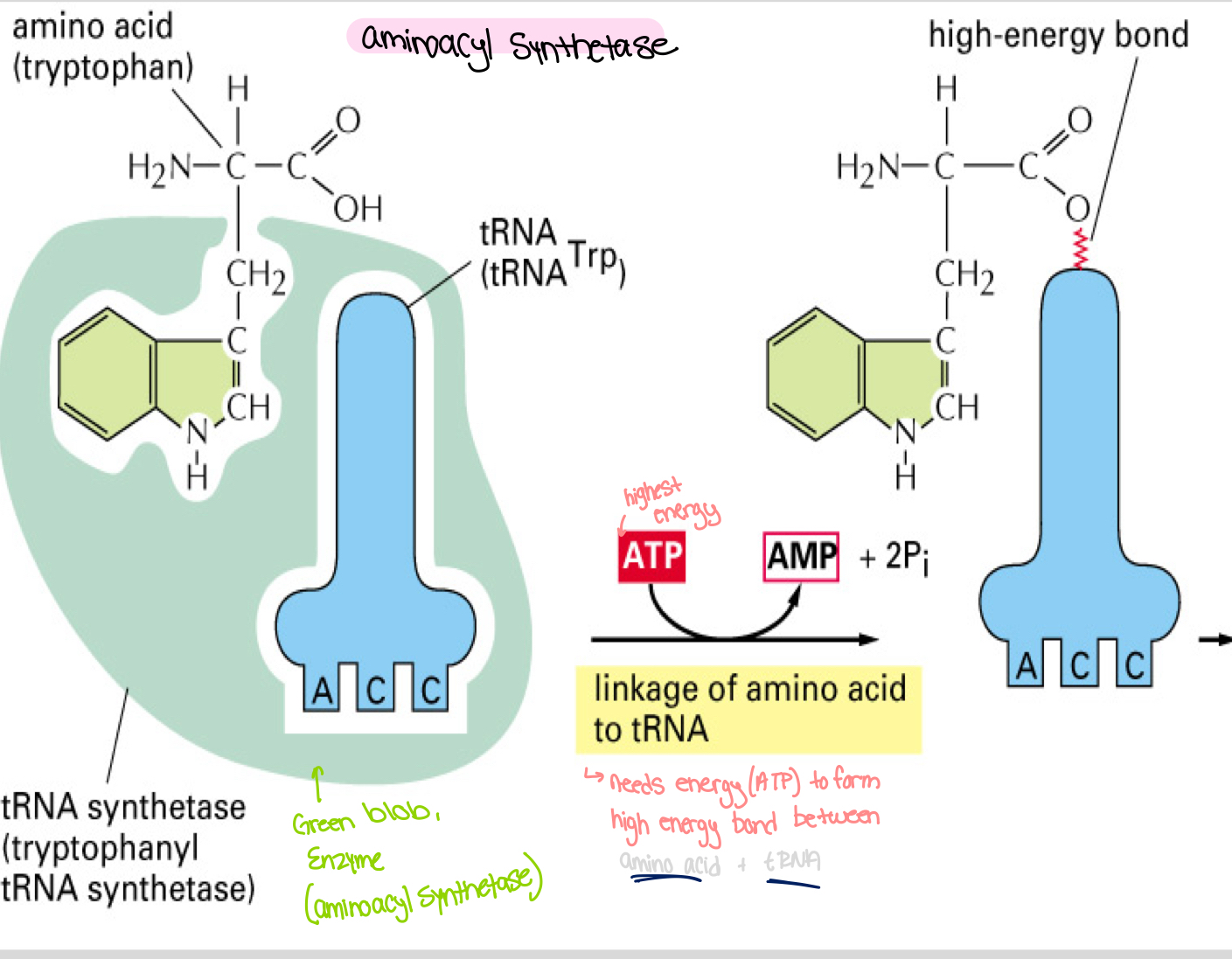
Aminoacyl Synthetase
Function: Enzyme that links an amino acid (example: tryptophan) to its corresponding tRNA.
Process: The attachment of an amino acid to tRNA involves the formation of a high-energy bond, utilizing ATP to convert into AMP and inorganic phosphates (2 Pi).
High energy bond will be used later to form peptide bond . The peptide bond formed will be lower energy, more stable .
tRNA Editing Mechanism
Editing Site: tRNA synthetase includes an editing site to remove incorrectly bound amino acids to ensure fidelity during synthesis.
Hydrolytic Editing: in some tRNA synthetase, mischarged amino acids can be hydrolyzed and removed, helping maintain accuracy in tRNA-amino acid pairing.
Ribosome Structure and Function
Composition: Ribosome is a large ribonucleoprotein complex made of two major subunits containing rRNA and over 50 proteins.
Eukaryotic Assembly: rRNA components are transcribed as pre-rRNA in the nucleolus and exported into the cytosol.
Millions of ribosomes exist in each eukaryotic cell.
Ribosome Organization
Small Subunit: Contains the mRNA binding site and tRNA binding sites.
Large Subunit: Catalyzes the formation of peptide bonds between amino acids.
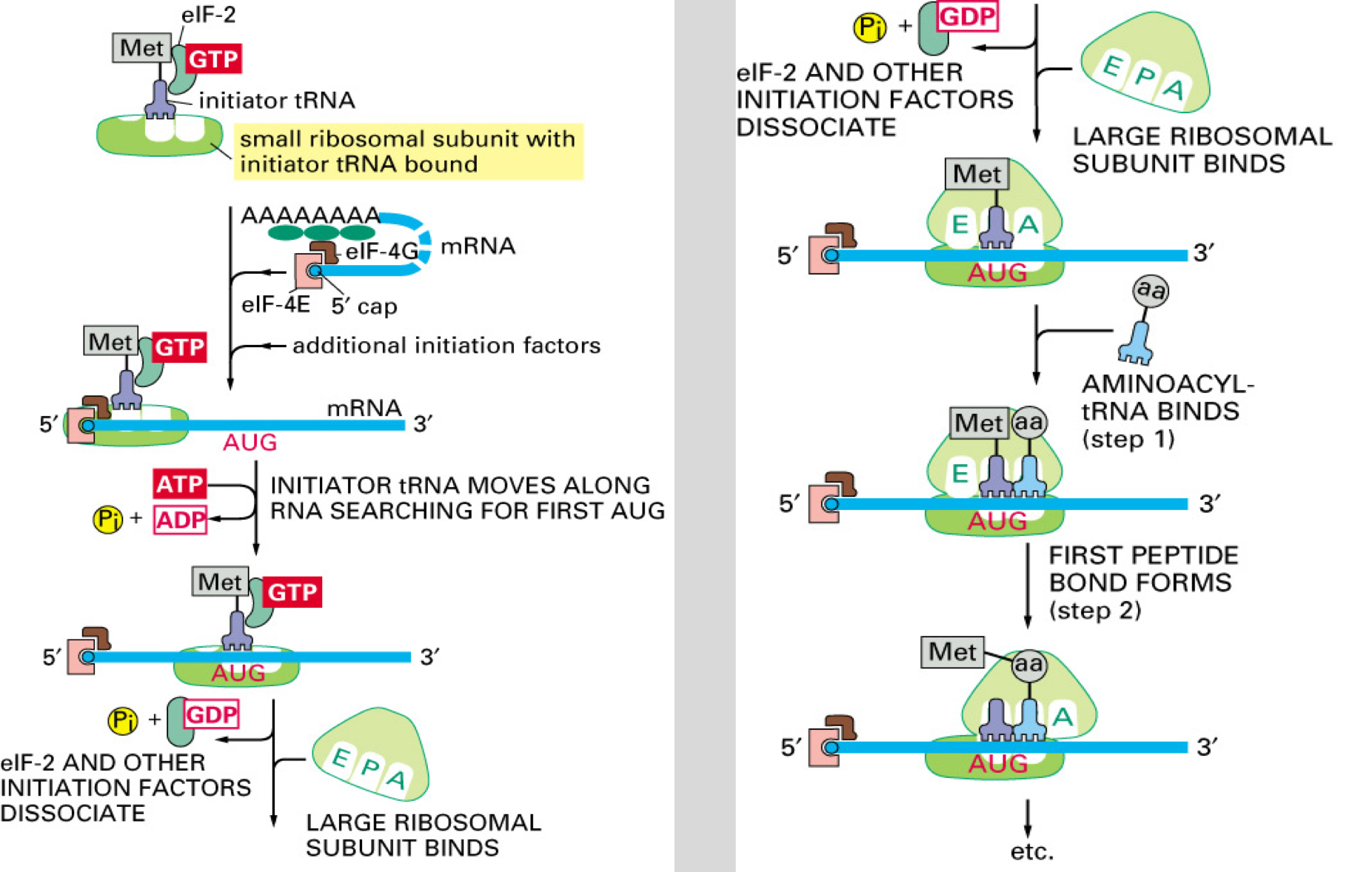
Eukaryotic Initiation of Translation
Initiator tRNA: The initiator aminoacyl tRNA loads onto the small ribosomal subunit (SRS).
Initiation Factors: Additional eukaryotic initiation factors (eIFs) bind to SRS, recognizing the 5' cap of mRNA and scanning for AUG start codon, powered by ATP hydrolysis.
Eukaryotic Start of Translation
Start Codon: Upon encountering AUG, eIFs dissociate; the large ribosomal subunit binds.
However : Early AUGs may sometimes be skipped in favor of a downstream AUG, a process known as 'leaky scanning.'
First AUG encountered reads as “start”
Bacterial Translation Start
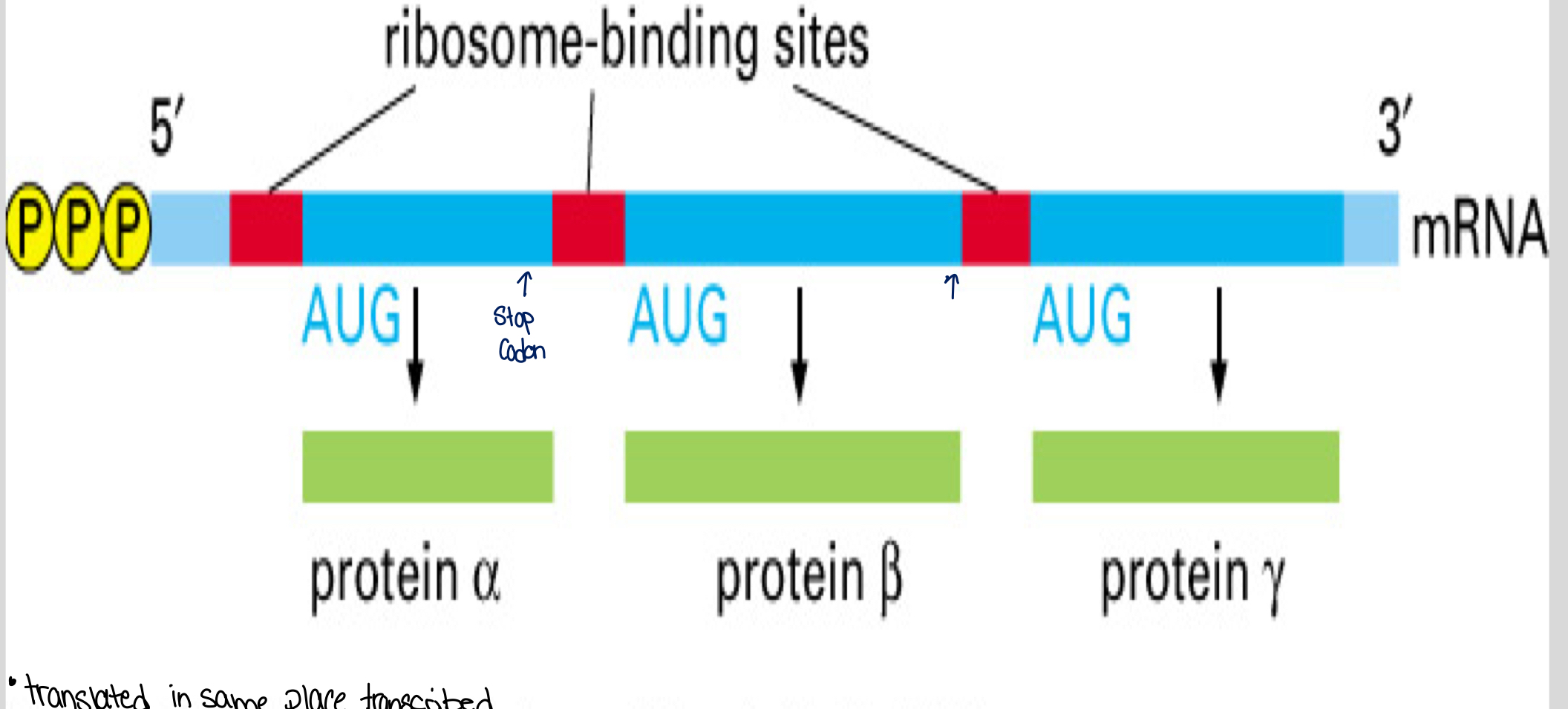
Consensus Sequences: Bacterial mRNAs lack a 5' cap; instead, they use consensus sequence signals for ribosome binding and scanning for their start codons (AUG).
Bacterial mRNA can be polycistronic, facilitating the synthesis of multiple proteins from a single mRNA strand.
Translation Elongation Steps
Step 1: The growing polypeptide chain elongates as incoming aminoacyl tRNAs enter the A site of the ribosome.
Step 2: A high-energy bond in the first amino acid is broken to form a new bond; this energy is used to catalytically form the peptide bond between amino acids.
Step 3: The large subunit shifts while the small subunit and tRNA remain stationary; the unloaded tRNA is then displaced to the E site.
Translation Termination
Stop Codons: UAA, UAG, and UGA act as signals for termination.
Release Factors: When stop codons are reached, release factors bind to the A site, adding water instead of an amino acid, thereby releasing the polypeptide from the tRNA and concluding synthesis.
Polyribosomes (Polysomes)
Definition: Many ribosomes translate the same mRNA strand simultaneously, enhancing protein synthesis efficiency.
Polyribosome Function: In bacteria, translation can start even before transcription completes; this is not possible in eukaryotic cells.
Efficiency of Translation
Trade-offs: Cellular processes like protein synthesis have a balance between accuracy and speed, generally favoring accuracy. High-energy bonds for each peptide bond formed reflect this trade-off.
Protein Folding
Folding Mechanism: Secondary and tertiary folding occurs as protein synthesis progresses.
Role of Chaperones: Molecular chaperones assist and sometimes accelerate tertiary protein folding, correcting minor flaws.
Types of Molecular Chaperones
hsp70 and hsp60: Major eukaryotic chaperones.
hsp70 binds to hydrophobic regions and, utilizing ATP, compels improperly folded proteins to unfold, allowing for proper refolding.
hsp60 forms a barrel-shaped structure, creating an isolated space for mis-folded proteins to refold correctly.
Quality Control and Proteasomes
Misfolded Proteins: Those proteins that cannot re-fold correctly by chaperones are targeted for degradation by proteasomes.
Ubiquitin Tags: A special class of protease degrades large peptides into smaller peptides; additionally, misfolded proteins receive ubiquitin attachments as a degradation signal.