L22: NMR spectroscopy II
Two-dimensional NMR
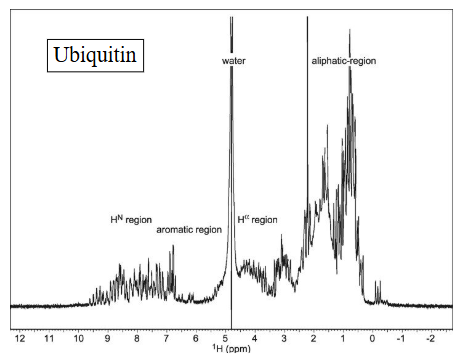
Protein 1D spectra are congested
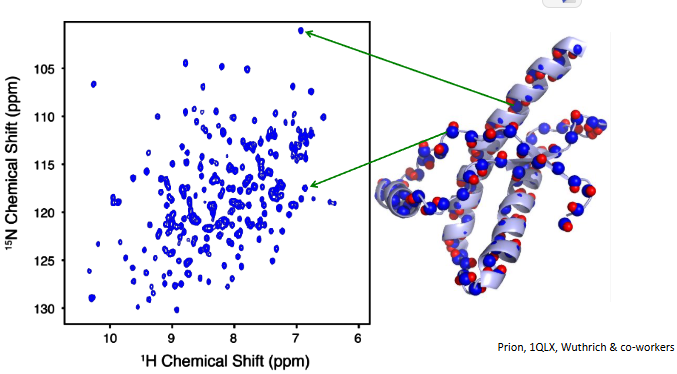
2D HSQC (Heteronuclear single quantum coherence) spectroscopy provides a fingerprint of a protein
establish correlation
two 1D experiments ≠ HSQC
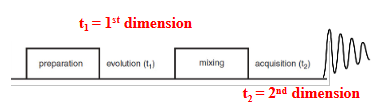
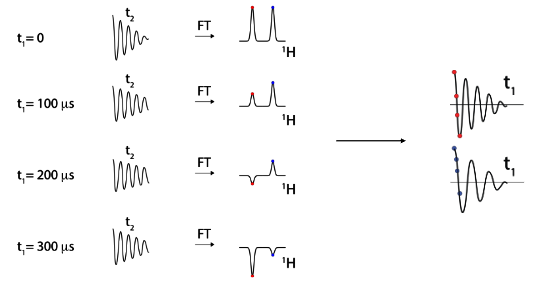
Preparation (pulses)
Evolution (chemical shift, 1st dimension)
Mixing (exchange magnetization)
Acquisition of FID (chemical shift, 2nd dimension)
HSQC pulse sequence
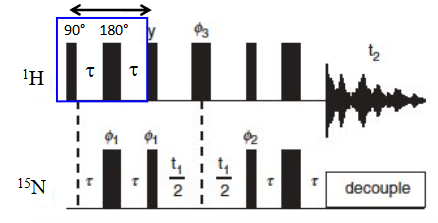
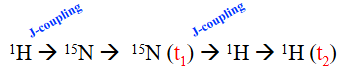
J-coupling is commonly used to create coherent states between nuclei for magnetization transfer.
time necessary for energy to transfer is related to J-coupling constant
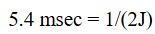
Hahn echo
90 → tau → 180 → tau → echo

Allows for magnetization transfer between nuclei without the occurrence of chemical shift evolution
can be used to enable “J-coupling evolution”
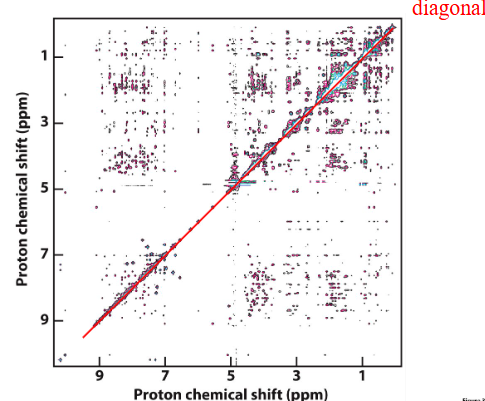
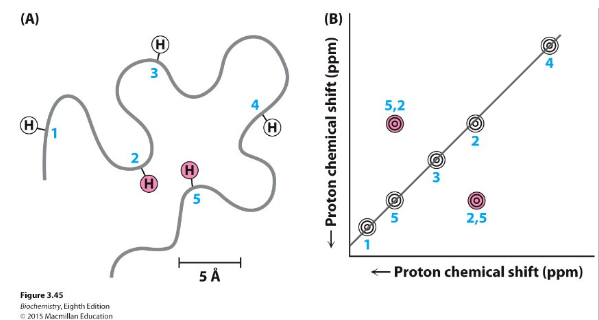
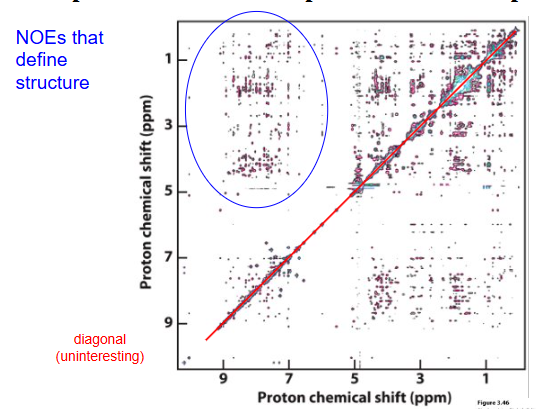
NOESY (Nuclear Overhauser Effect SpectroscopY)
normally measured between protons
Exchange of magnetization CROSS RELAXATION if within ~5 Å (during “mix time”) - carries information from the other when they ‘talk’
t1 = 0 first time, then increases
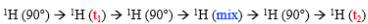
Schematic
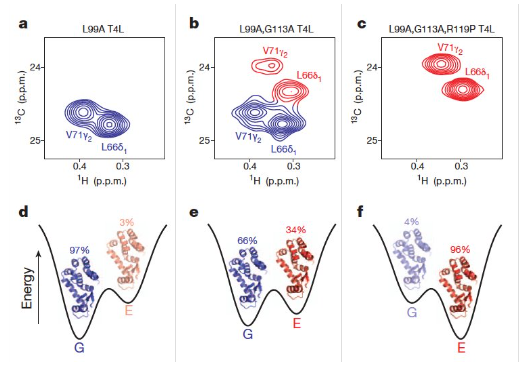
Chemical shift perturbation (CSP) approach
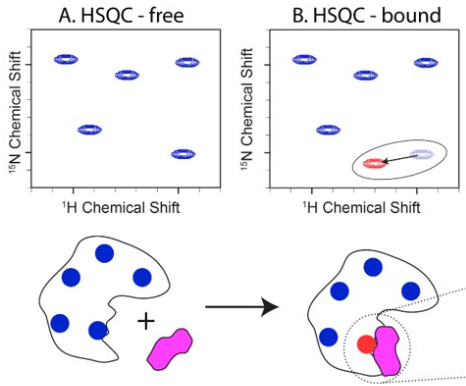
Assumption: chemical shift changes are close to binding site
SAR by NMR
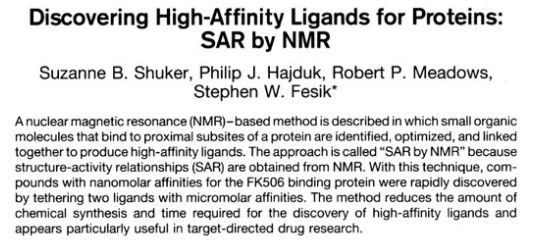
Unique approach to drug discovery that aims to find high affinity ligands using lower affinity fragments that would otherwise be overlooked in screening assays.
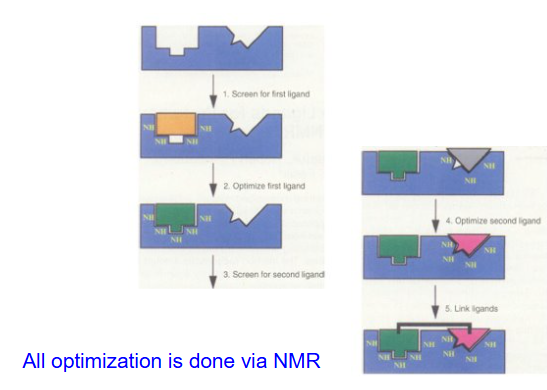
improve poor binders
Binding affinities change from ~1 mM to ~40 nM (factor of 25,000X)
Structure determination approach
not first choice… but preferred with small and disordered targets
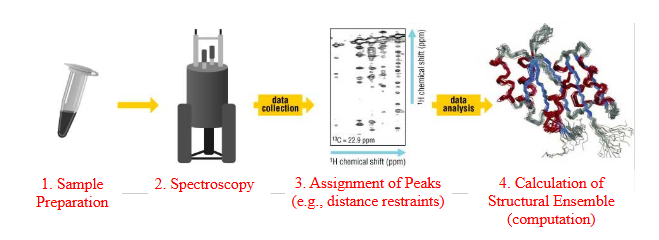
1. Sample Preparation (incorporation of NMR labels)
2. Acquisition of NMR data
3. Assignment of NMR spectra (NOESY, etc.)
4. Calculation of structure with NMR restraints
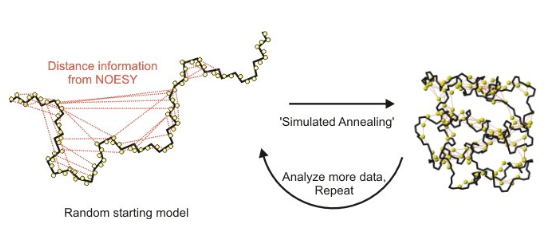
metallurgy :)
Restraints needed for NMR structure
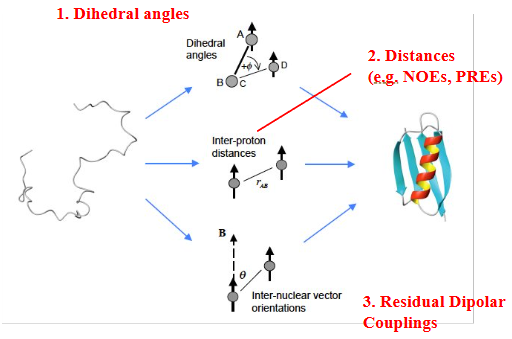
Dynamics by NMR
Protein motion and NMR
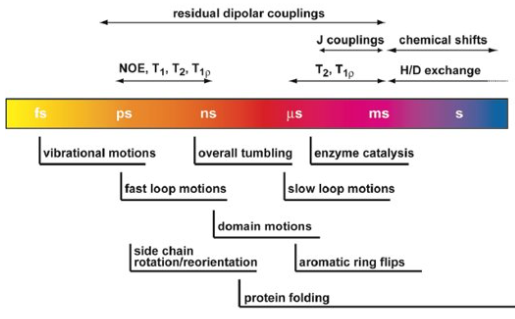
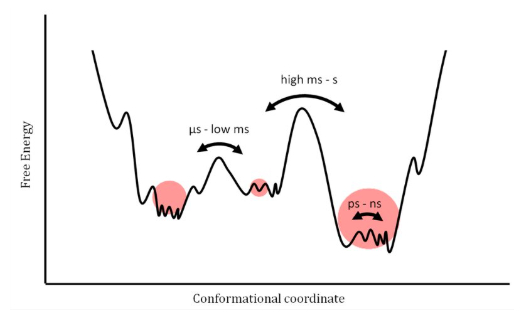
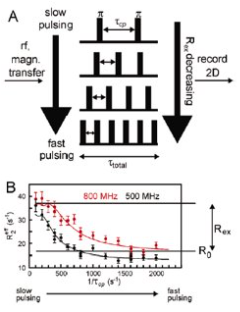
conformational exchange (slow - ms-s)
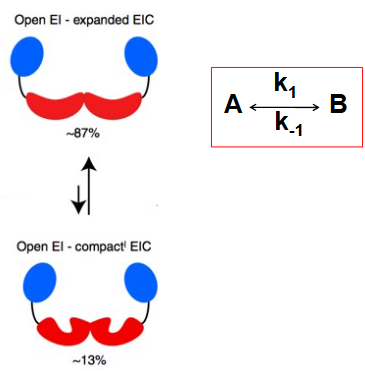
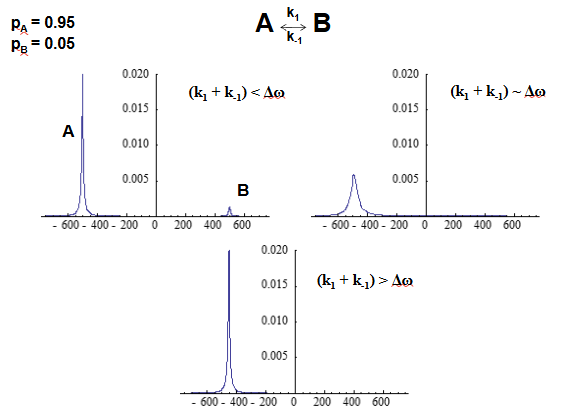
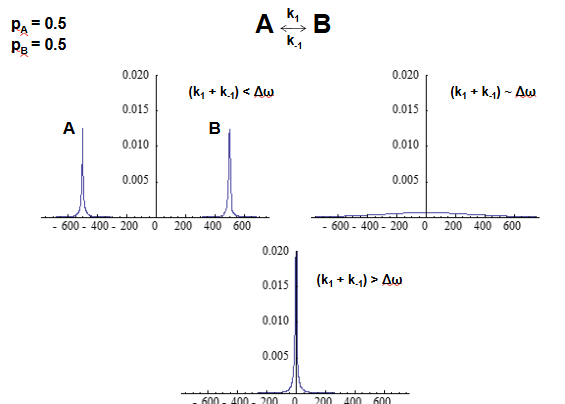
CPMG relaxation dispersion experiment
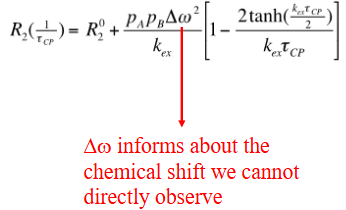
chemical shifts are sensitive to secondary structures
spin relaxation (fast - ns)
Relaxation drives spins back to their equilibrium distribution
Relaxation is a natural phenomenon driven by molecular motions.
(i.e., relaxation is caused by random, fluctuating magnetic fields around the sites of nuclear spins - disorder of magnetization vectors)
Spin relaxation experiments can be used to study molecular dynamics/motions, which is one of the greatest strengths of NMR
Longitudinal relaxation - The return to the z axis is described by T1 (1/R1)
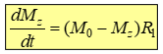
Transverse relaxation - Loss of x/y coherence is described by T2 (1/R2)
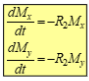
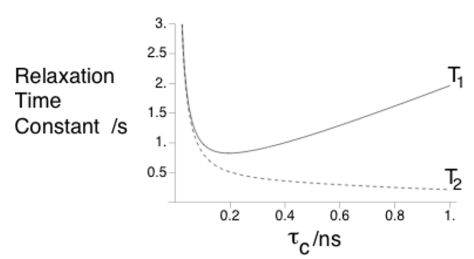
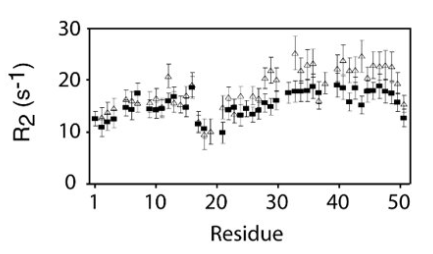
R2 increases (decrease in T2) is indicative of longer local correlation time
Transverse magnetization
When the net magnetization is in the x/y plane
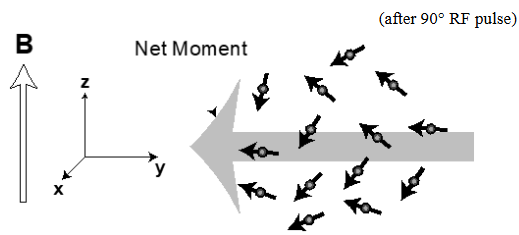
Transverse magnetization decays due to microscopic differences of the magnetic fields surrounding each nuclear spin in the ensemble - loss of coherence
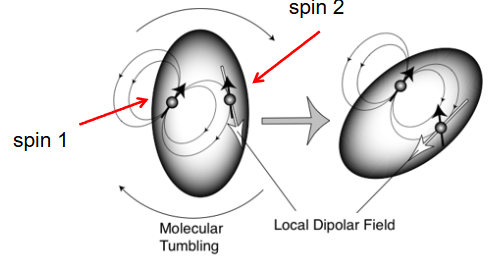
Dipolar field fluctuations and molecular tumbling
Reorientation of molecules relative to B0 gives rise to time dependent local magnetic field fluctuations
Correlation time: Stokes law for Brownian rotational diffusion
time it takes on average to rotate by pi radians
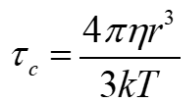
r is the radius
η is the solvent viscosity
k is the Boltzmann constant
T is the temperature
TAKE HOME
Solution NMR can be carried out on all size proteins.
False! Intrinsic limitation in relaxation coherence