U4AOS2 how are species related over time?
selection pressures need to added
flash cards are done, may need to add more idk
#daffe7 - highlighter colour Green
#fcf4c5 - Highlighter colour yellow
Mutations occur in the sex cells, after meiosis of a gamete (eggs + sperm), transferring into offspring.
Variation
Variation in the phenotypes of individuals in a populationresults from the different combinations of alleles (genotypes) that have arisen due to sexual reproduction and meiosis, and the influence of the environment.
For example, a plant may have a genotype that allows it to grow tall, but it will only be able to do so if it has access to all the requirements for growth.
→ If these requirements are lacking, then the plant will not grow to the size determined by its genotype.
→ The environment has influenced the phenotype.
Types of Variation possible:
→ Morphological: differences in the structure of an organism; for example, body shape, tail length in dogs
→ Behavioural: differences in patterns of activity; for example dog behaviour (herding, retrieval, guarding)
→ Biochemical: differences in the composition of cells; for example, blood groups and the different pigments in cat hair
→ Physiological: differences in the functioning of an organism; for example, differences in people’s ability to detect tastes, odours and colours
→ Developmental: differences that occur as an organism ages; for example, a snake changing colour over time
→ Geographic: differences in location; for example, different possum body mass in different regions of Australia.
A species is a group of organisms that can interbreed and produce fertile and viable offspring in their natural environment
Gene Pools
A gene pool is the sum total of alleles present in a population of organisms.
We can use gene pools to determine allele frequency, which is the proportion of a particular allele within a population.
The range of variations possible in a population is limited by the alleles that are available in the gene pool.
In a large gene pool, there is likely to be a greater number of different alleles, and therefore more genetic diversity,
while in a small gene pool, there is likely to be less genetic diversity.
Mutations
When Changes in the DNA is permanent, its called a mutation.
The result can change the nucleotide sequence of a section of DNA, therefore affecting the protein produced
New alleles arise from this process, causing variation in individuals
New alleles mean there will be changes in the allele frequencies of a population over time and this, you will recall, is the definition of evolution.
Mutations that occur in genetics can be passed on to offspring.
Mutations can occur naturally or be induced.
→ Spontaneous mutations are naturally occurring changes caused by errors in DNA replication. They create new alleles instantly.
→ Induced mutations are caused by exposure to external elements. Induced mutations alter DNA at a slower rate than naturally occurring mutations.
there are two Categories of mutation:
Gene Mutations
Block (or Chromosomal) Mutations
Gene Mutations
Gene Mutations can include Point Mutations or Frameshift Mutations
Point Mutations
A small, localised change to one base in the nucleotide sequence of a gene:
WHEN SUBSTITUTING A BASE IN SEQUENCE
silent mutations: one nucleotide is replaced with another that codes for the same amino acid
missense mutations: one nucleotide is substituted for another resulting in a different amino acid being coded for.
nonsense mutations: where one nucleotide is substituted for another resulting in a stop codon, no amino acids are produced after this point.
Frameshift Mutations
Where one or more nucleotides are inserted or deleted from the DNA which results in the reading frame of the DNA to change
Will alter the codon reading frame and therefore affects every codon, and therefore every amino acid, beyond the point (downstream) of the mutation.
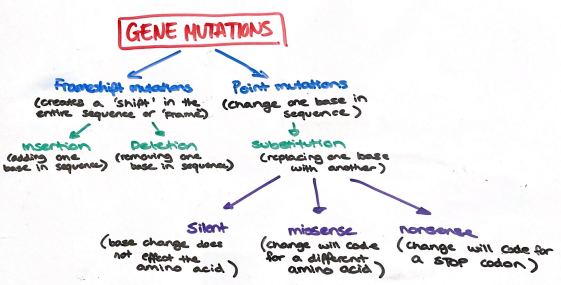
** silent, missense and nonsense mutations are the results of deletion, substitution and addition.
allele frequency: is a change in the nucleotide sequence of a section of DNA. new alleles are formed by mutation, they can also be:
beneficial mutations to the gene creating new variations of a trait (missense mutations).
detrimental truncate (cut) gene sequence to abrogate the normal function of a trait
Gene mutations can be beneficial, detrimental or neutral (nonsense mutations).neutral mutations have no effect (silent mutations).
chromosomal abnormalities (aneuploidy): non-disjunction refers to the chromosomes failing to separate correctly, resulting in gametes with one extra, or missing, chromosomes.
if a zygote is formed a gamete that has experiences a non-disjunction event, the resulting offspring will have an extra or missing chromosome in every cell of the body.
e.g
Patau syndrome (trisomy 13)
Edwards syndrome (trisomy 18)
down syndrome (trisomy 21)
klinefelter syndrome (XXY)
turners syndrome / fragile x (monosomy X)
Compare aneuploidy and polyploidy. (4-5 marks)
Block Mutations
Block mutations are also called chromosome mutations, as they alter large sections of DNA containing multiple genes.
→ Such mutations are usually a consequence of spontaneous errors during meiosis.
There are several types of block mutations and each can have significant effects on an organism’s phenotype, depending on the location of the change.
→ For example, genes may be disrupted, removed or disabled.
DELETION: A section of DNA is removed from the chromosome; Leads to disrupted and/or missing genes; Large impact on growth and development, sometimes fatal.
DUPLICATION: A section of chromosome is repeated multiple times (sometimes thousands of times) on the same chromosome; Can lead to an increase in gene expression.
INVERSION: A section of chromosome is removed, rotated 180° and reattached in reverse order; Can be as small as two bases, or as large a several genes; Usually does not cause abnormalities, but can cause lowered fertility.
TRANSLOCATION: A section of one chromosome attaches to a non-homologous chromosome or sections are exchanged between the two; Leads to disruptions in gene sequences or interruptions in gene regulation; Can cause some cancers
Adaptations
definition: is a trait that allows an organism to outcompete all other organisms in a species, allowing for a longer lifespan to produce offspring. mutation, selective advantage
natural selection of a single variation, that does not generally bring about much change. first variation is selected, then individuals with it develop another. second variation is selected, then a third, ect.
many new variations also are not selected, competing for nutrition, mates and not get eaten. as the process continues, organisms have traits that make them very well suited tot heir environments. developing new ways of hiding, hunting, protecting, keeping cool and warm, avoiding pests to increase chances of survival and reproduction. These are adaptations.
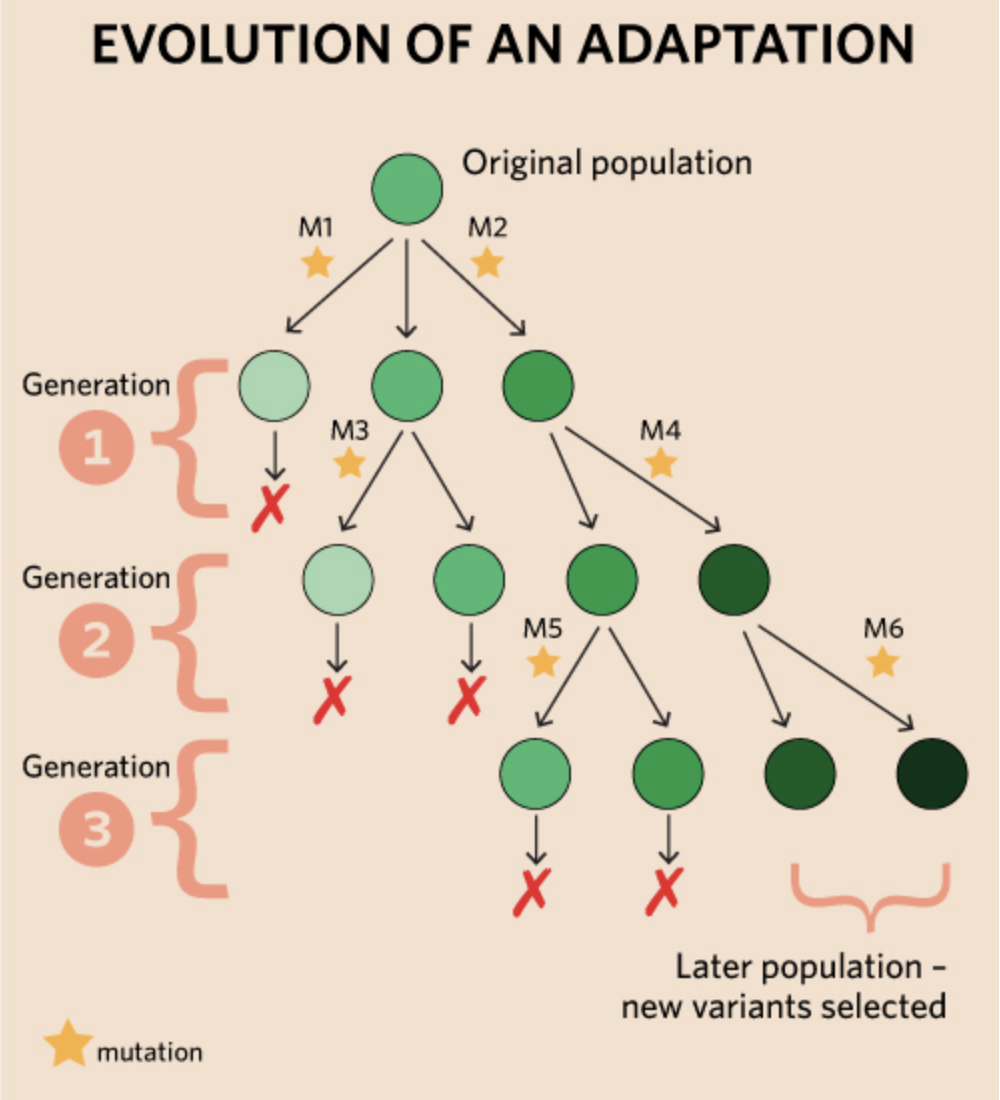
Species: evolution also explains how different species can come from a single ancestor species.
a species are populations that breed amongst themselves. often species don’t intermix, but it can occur.
e.g black and brown bears
their territories overlap but they don’t always mate, but if they do they can produce healthy offspring.
Gene flow and drift
Gene flow: is the exchange of genetic information, specifically alleles, into or out of a population of the same species.
In animals, gene flow results from their movement into (immigration) or out of (emigration) a population.
In plants, it results from the movement of seeds and pollen.
populations gain or lose alleles though gene flow
immigration - new individuals enter the population
emigration - leave population
When genes ‘flow’ from one population to another, that flow may increase genetic diversity of the gene pool of the receiving population, affecting the allele frequency.
Gene flow reduce the differences between populations (interbreed and genes shared/transferred). this makes gene pools more similar.
But what impact does it have?
have a greater effect on smaller populations, some may only carry a particular gene and if they leave the population it can be lost. In larger populations it is likely that some other individuals will still possess the same, same for immigration. therefore gene flow can result in the loss or introduction of new alleles.
Gene drift: a random change in allele frequency, occurring naturally in every population, due to chance events.
In genetic drift, no allele is favoured; they are all equally subject to being affected.
In small populations, genetic drift can have a much more dramatic effect than in large populations, as it can lead to a reduction in genetic diversity.
As time progresses, alleles may be lost from the gene pool (frequency is 0%) or fixed as the only allele present for the gene (frequency is 100%).
Events in the environment may ‘magnify’ genetic drift, by randomly killing a part of the population. Such events may be volcanic eruptions, landslides, floods and bushfires – but they also include the impact of disease, predators and so on.
eg. Take the population of goldfish in a bowl, where the frequency of the two alleles present is very different (two DD, four Dd). Suppose a cat randomly catches two fish, which happen to both have the genotype DD. The frequency of each allele left in the population, D and d, is now equal. As the fish reproduce from now on, there will be a change in the allele frequencies of the population over time, meaning that evolution is occurring.
There are two effects of genetic drift that may be severe:
Bottleneck effect
Founder effect
Bottleneck effect
Bottle neck effect: population bottlenecks occur when an event reduced population size by an order of magnitude (>50%)
- Bottlenecks may result from natural occurrences (fires, floods, etc) or be human induced (overhunting).
- The reduction in genetic diversity causes changes in the allele frequency of the gene pool, and thus the population is evolving.
- The population may recover and increase its numbers again, having squeezed through a ‘bottleneck’ of low numbers.
- Surviving population has less genetic variability then before and will be subject to a higher level of genetic drift
- However, as the surviving members of the species begin to repopulate, the allele frequencies of the new population are no longer representative of the original gene pool.
- Alleles may be lost entirely after one natural disaster or may disappear after several generations of reproducing.
e.g. Northern elephant seals have reduced genetic diversity compared with southern seals due to overhunting.
- Founder effect occurs when a small group breaks away from larger population to colonise a new territory
- the population subset does not have the same degree of diversity as a larger population, subject to more genetic drift
new colony increases in size, its gene pool will no longer be representative of the original gene pool
founder effect differs from population bottleneck in that the original population remains largely intact
e.g. certain amish communities have higher incidence of polydactyly because of inter-marriage within the community
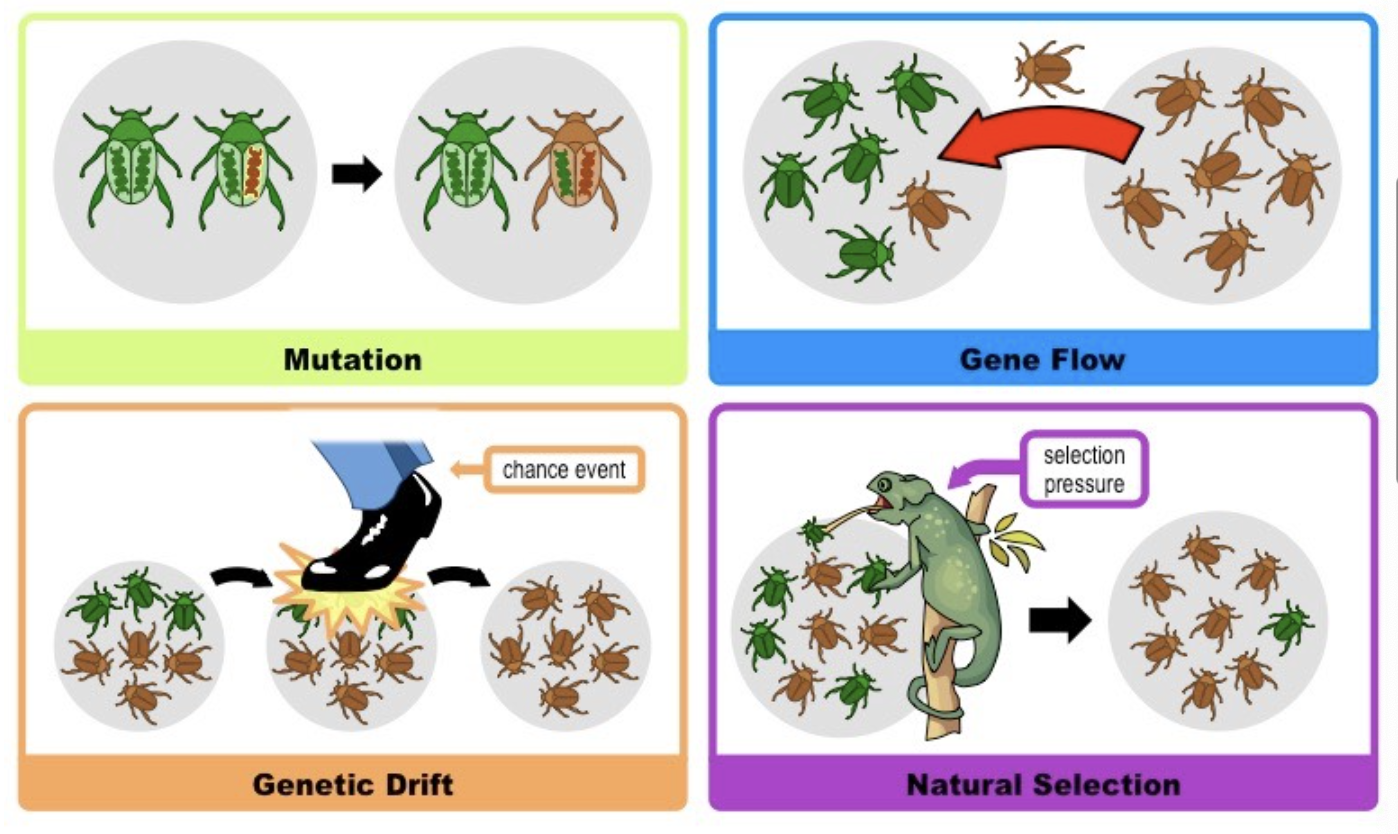
Artificial selection
nature - chooses selections pressures. artificial - humans are the selection pressure
Humans select which traits are beneficial and breed organisms to produce these traits. Faster evolutionary change as humans discard undesirable traits to prevent them from remaining in the population. Resulting traits are not necessarily beneficial for the organism but are desirable for humans. driven by human intervention:
what looks good (aesthetics)
monetary drive
Problems with selective breeding: ethics question
Future generations of selectively bred plants and animals will all share similar genes. Making some diseases more dangerous as all the organisms would be affected. Also, there is an increased risk of genetic disease caused by recessive alleles.
All the genes and their different alleles within a population is its gene pool. Inbreeding can lead to the loss of alleles from the gene pool, making it more difficult to produce new varieties in the future.
four steps involved in artificial selection and how do these differ from natural selection?
variation
intervention
heritability
breeding
step three is different - instead of selection pressures in natural selection, intervention by humans replaces this step
Pathogen Evolution
bacteria: antibiotic use (selection pressure), resulting in antibiotic resistance when antibiotics are not used correctly or in overuse. Evolution occurs when the majority of a species of bacteria gain the antibiotic resistance gene.
viruses: very fast evolution due to high reproductive rate. antigenic drift; point mutations, proteins on surface change, antibodies to pervious versions of virus will not attach, therefore epidemics occur, this occurs frequently up to multiple times a year. antigenic shift; two viruses infect a cell, their genetic material recombines to form a totally new virus, no immunity anywhere in the world, thereby resulting in a pandemic if it is highly contagious, it is a rare event (10 in last 100 years)
Speciation
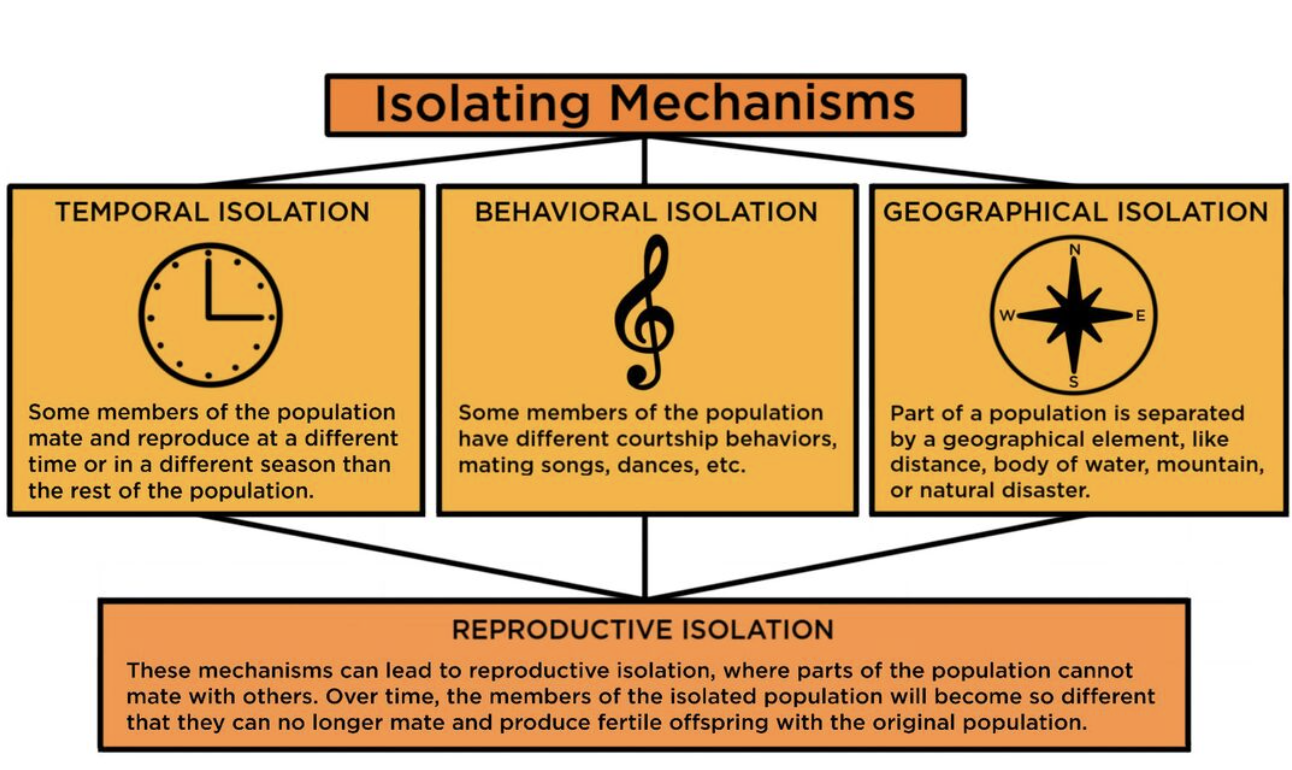
the process where new species are formed due to differing selection pressures. Allopatric speciation (divergent): geographic isolation of species.
a population of organisms become geographically isolated
each group will undergo different selection pressures where they will undergo natural selection/genetic drift
after many generations, the two populations have accumulated many adaptions/mutations where they become reproductively isolated.
when the two populations are joined, they will not mate, therefore speciation has occurred.
sympatric speciation: live in the same geographical area.
→ temporal isolation (timing) - flowering different times, mate at different times
→ behavioural isolation (cannot communicate) - doesn’t recognise mating rituals, or language
→ genetic variation (post zygotic changes) - gamete mortality, zygote mortality, sterility of offspring.
Fossils
preserved evidence of organisms
body fossils: plant/animal/algae, and preserved remains (fossil formation, frozen, tripped in amber, preserved in bog.
trace fossils: evidence that organism was alive (poo, foot prints, tools)
dating fossils: geologists often need to know the age of the material they find. they use absolute dating methods, sometimes called numerical or radiometric dating, to give rocks an actual date or date range, in numbers of years. this is different to relative dating which only puts geological events in time order.
absolute dating (using radioactive elements): to understand it is necessary to understand the atom. the nucleus of an atom is made up of protons and neutrons, the number of protons define the type of element. however, the number of neutrons may vary, if an atom with same number of protons and different neutrons are called isotopes.
isotopes are stable but some can be unstable. they can undergo a process called radioactive decay, where they spontaneously change to elements to a different type. it can never be predicted when a specific atom will undergo radioactive decay. when considering atoms its seen that the decay occurs at an exponential rate, meaning that over a certain period (half life), half the unstable isotopes will undergo radioactive decay. radiocarbon dating is the most useful radiometric dating methods.
there are 3 types of rock found on earth:
sedimentary - small pieces of rock that are stuck together with mineral glue
igneous - from volcanoes
metamorphic - sedimentary or igneous rock that has had lots of heat/pressure applied over time to change it
relative dating: is the method of determining the order of events from the fossil record. by studying the order in which fossils occur in the fossil record, geologist can determine the order of events as they occurred but not when exactly they occurred. Fossils found at teh lowest layer of rocks would be the oldest, as these would have been buried for the longest time, whereas fossils found closer to the surface would be buried more recently and therefore be younger. the geological time-scale studied was entirely developed by relative dating methods. it is useful method of dating when fossil materials lack radioactive isotopes.
index fossils: when the bones of our early ancestors are found in the same geological strata as those of other animals that are known to have lived only during a specific time period in the past, we assume that these ancestor must also have come from that time. this is referred to as dating by association with index fossils since they underwent relatively rapid evolutionary changes that are identifiable in their teeth and other skeletal parts. their bones also were frequently found in association with our human and primate ancestors
Defining human
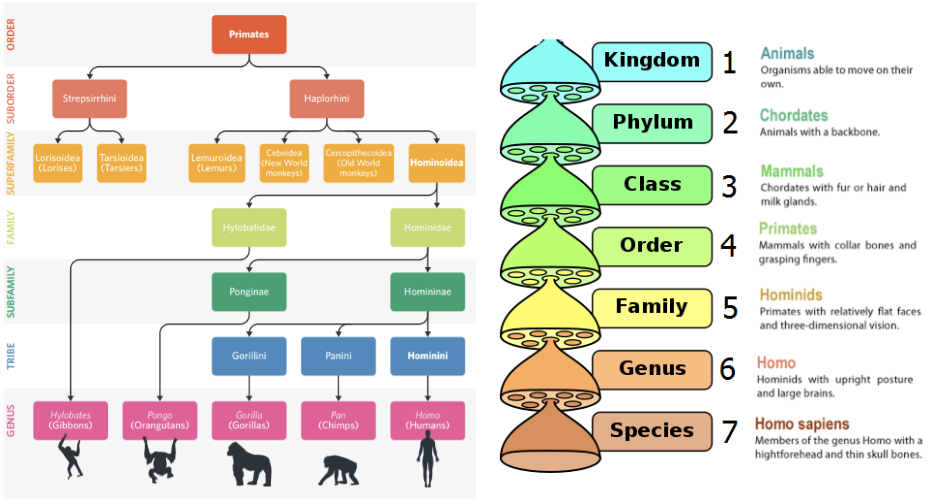
The four features that define characteristics of mammals are?
Production of milk (mammary glands) to feed young
Fur/hair on the body
One lower jawbone
3 bones in the inner ear
Main features to focus on: Prehensile hands/feet with five digits with an opposable digit (either the thumb in the hand or the hallux in the foot) which allows us to have power in our grasp.
many touch receptors in fingertips
Flexible spines and a large degree of rotation (hips and shoulders) for living in trees
Large cranium with increased brain size
3D colour vision and forward-facing eyes which gives spatial awareness.
Live in groups as they are social animals
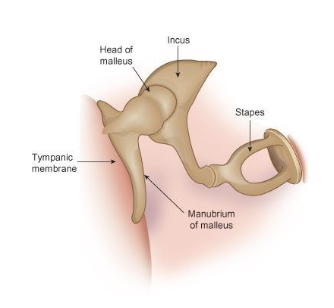
It is the inside structure of the ear, the three main bones which allow for better hearing, allowing for better transfer of vibrations from eardrum to inner ear.
hominoid: any great ape (such as humans) belonging to the superfamily Hominidae
Hominid – the group consisting of all modern and extinct Great Apes (that is, modern humans, chimpanzees, gorillas and orangutans plus all their immediate ancestors).
Hominoids tend to have the following features:
• Body features: Shorter spine between the rib cage and pelvis – this helps hominoids sit upright. Lack of tail – further contributes to hominoids ability to sit upright. Broader rib cage and pelvis – these also help hominoids sit upright. Typically, longer arms than legs and shoulder blades that sit further back give hominoids the ability to use their arms in many ways. Homo sapiens, however, are an exception to this and don’t have longer arms than legs.
• Skull features: Distinctive molar teeth in lower jaw – teeth that have five cusps arranged in a 'Y5' pattern. Increased cranium size – hominoid brains tend to be larger than other primates.
Hominin – the group consisting of modern humans, extinct human species and all our immediate ancestors (including members of the genera Homo, Australopithecus, Paranthropus and Ardipithecus).
3 distinguishing characteristics
• Bipedalism
• Communication and formation of complex social groups
• Structural consequences of bipedalism – centralised foramen magnum, S–shaped spine, broader rib cage, bowl–shaped pelvis, increased carrying angle of femur.
classification of humans:
kindgom - animal
phylum - chardata
class - mammals
order - primate
family - hominid
genus - homo
species - sapien
primates share these traits:
binocular vision and 3D vision
dextrous hands (some have opposable thumb)
tail/no tail (hominids dont have tail)
quadrupedal (4 legs) all other primates. bipedal (2 legs) - genus homo
complex brain - social development, communication, problem solving
flexible spine
Evidence of evolution
structural evolution: structural evolutionary changes over time as evidence of evolution. there are two types:
divergent (1 dividing into 2): evolution of species from a recent common ancestor. organisms subject to different selection pressures develop different structures, internal body chemistries and behaviours over time. this causes them to diverge from their parent species. adaptive radiation (1 dividing into many) is a type of divergent evolution where a single species rapidly diversifies into multiple species. can cause massive increases in biodiversity and occurs in ecosystems with lots of unfilled niches.
convergent: this pattern of evolution describes the process by which species from different parent species come to resemble each other because they have similar ecological roles, and natural selection has shaped similar adaptions.

Do you think a shark and a dolphin have a similar streamline shape due to divergent or convergent patterns of evolution? Explain your response. They do not have a recent common ancestor but have similar features due to selection pressures in their environment. thereby they do not have similar shape because of patterns of evolution.
structural morphology - as evidence of evolution
structural morphology: is the study of the form and structure of organisms. the field of comparing the structure of organisms is referred to as comparative anatomy. if organisms are related by evolution from a common ancestor, it would be expected that they would show similarities in their structural features, regardless of way of life.
homologous structures (divergent evolution): organs or skeletal elements of animals and organisms that, they virtue of their similarity, suggest their connection to a common ancestor. these structures do not have to look exactly the same, or have same function. they have similar structure but different functions. this is resulting from mutations and natural selections
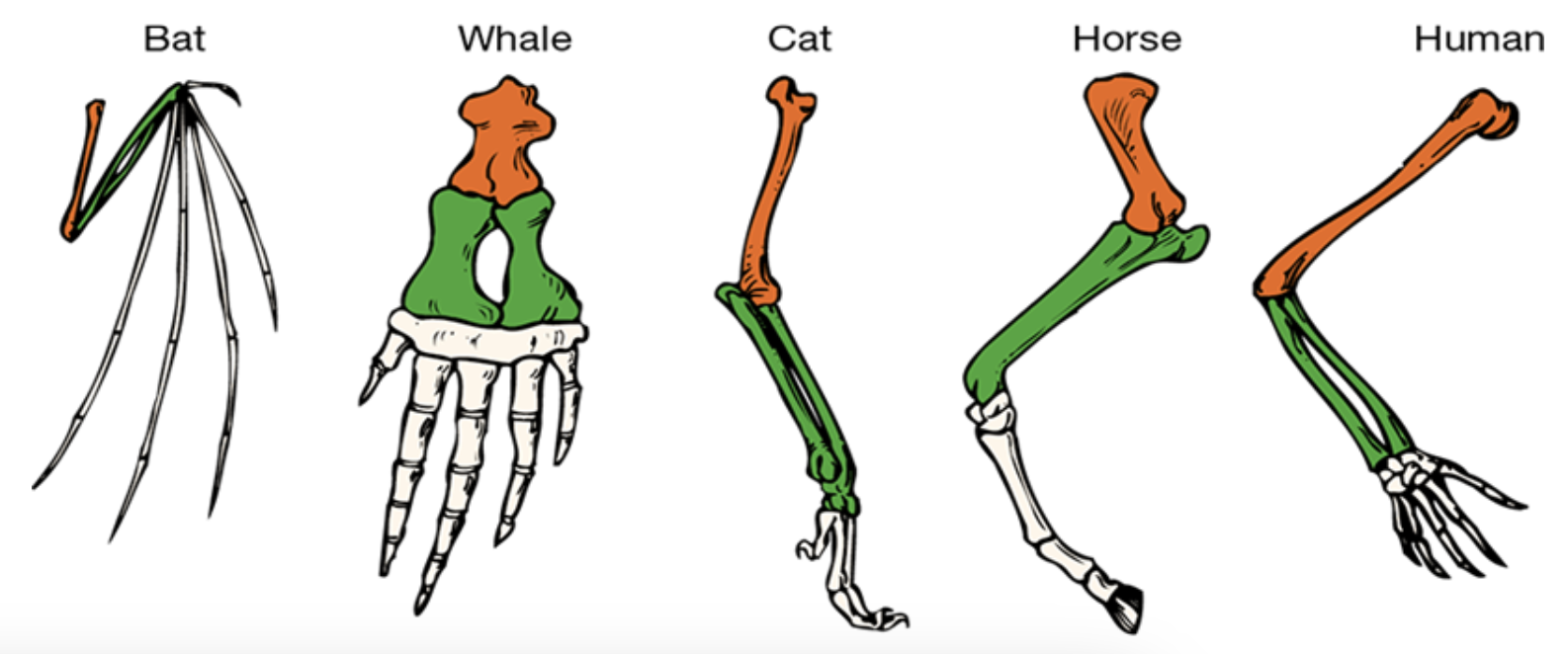
they all have similar bones but the ‘fingers’ have evolved and changes due to mutations and natural selections. e.g. the house has all fused together because they are walking on hard surfaces.
analogous structures (convergent evolution): comes from the root word “analogy” where two different things are compared based on them being alike in some way. analogous structures are similar structures that evolved independently in two living organism to serve the same purpose. they arise, not from a common ancestor, but from the organisms having to adapt to similar selective pressures
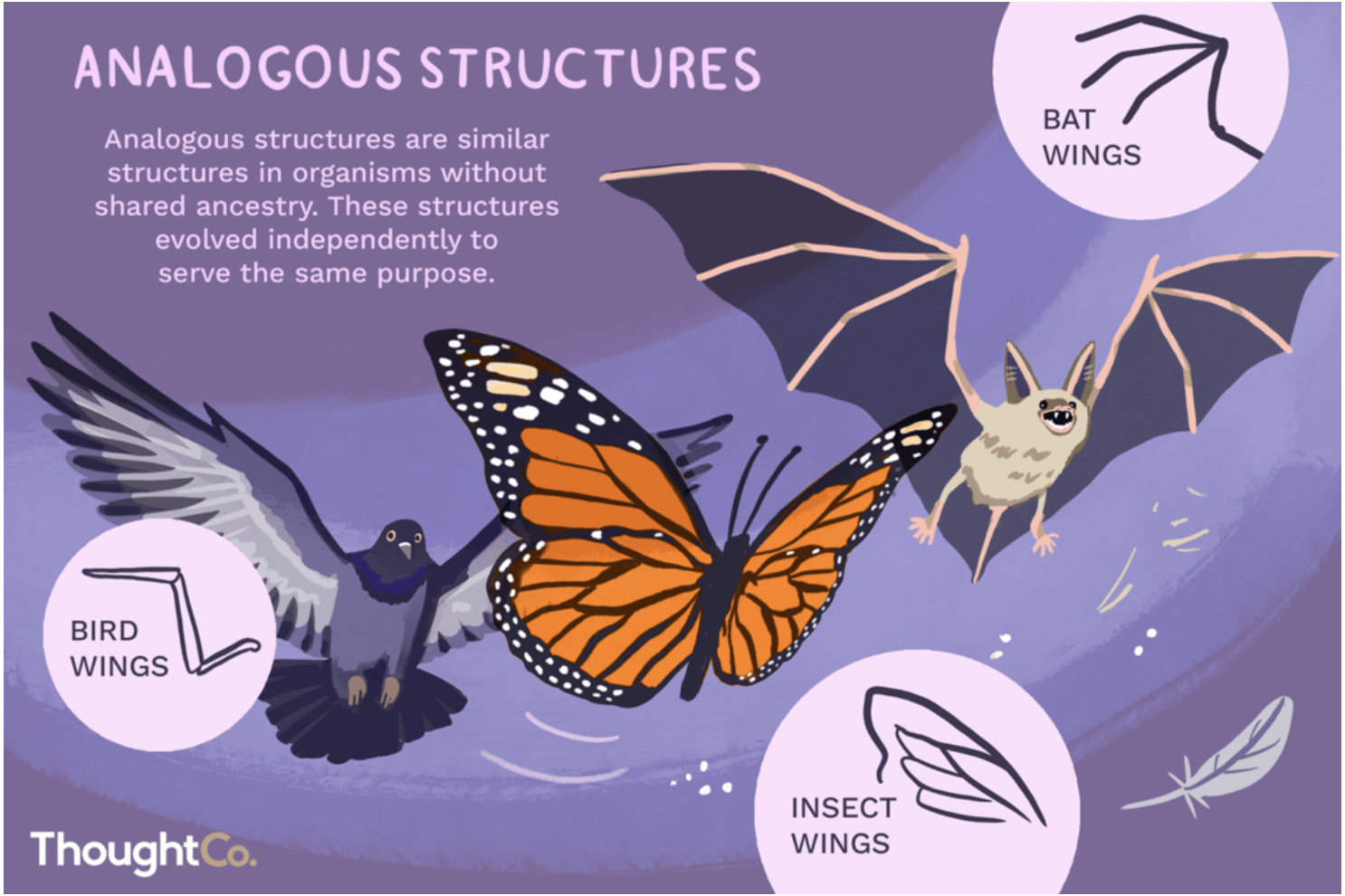
vestigial structures: often homologous to structures that function normally in other species. the existence of vestigial traits can be attributed to changes in the environment and behaviour patterns of the organism in question. as the function of the trait is no longer beneficial for survival, the likelihood that offspring will inherit the ‘normal’ form of it decreases.
determining relatedness - molecular homology
tools used to determine evolutionary relationships: fossils, morphology (homologous structures, and molecular evidence (DNA and amino acids).
molecular homology is used to determine relatedness between species using DNA and amino acids. as two species diverge from a common ancestor, they accumulate different mutations in their DNA.
Can look at how they are expressed in comparing common gene and amino genes and their nucleotide sequences, comparing whole genomes, comparing mitochondrial DNA.
amino acids - the degree of similarity in the amino acid sequence of the same protein in two species will, therefore, determine how closely related they are.
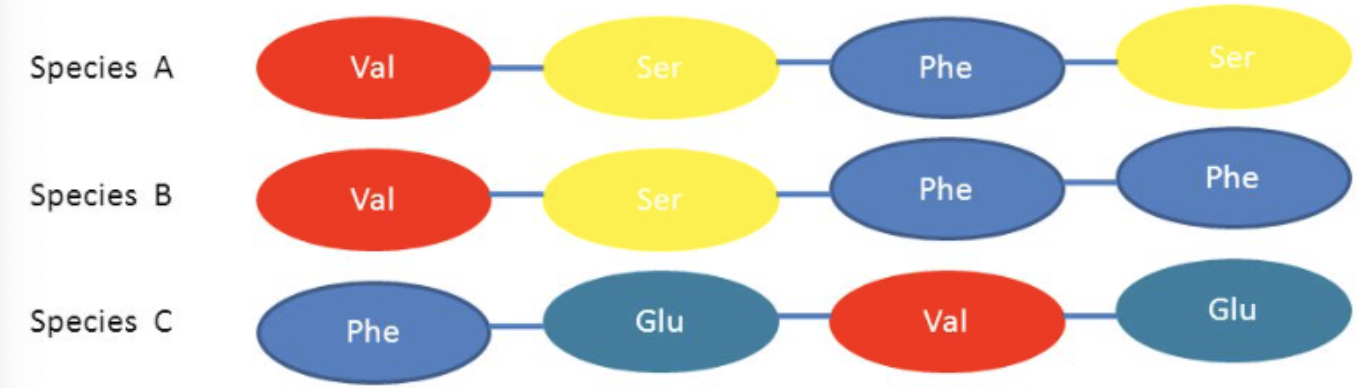
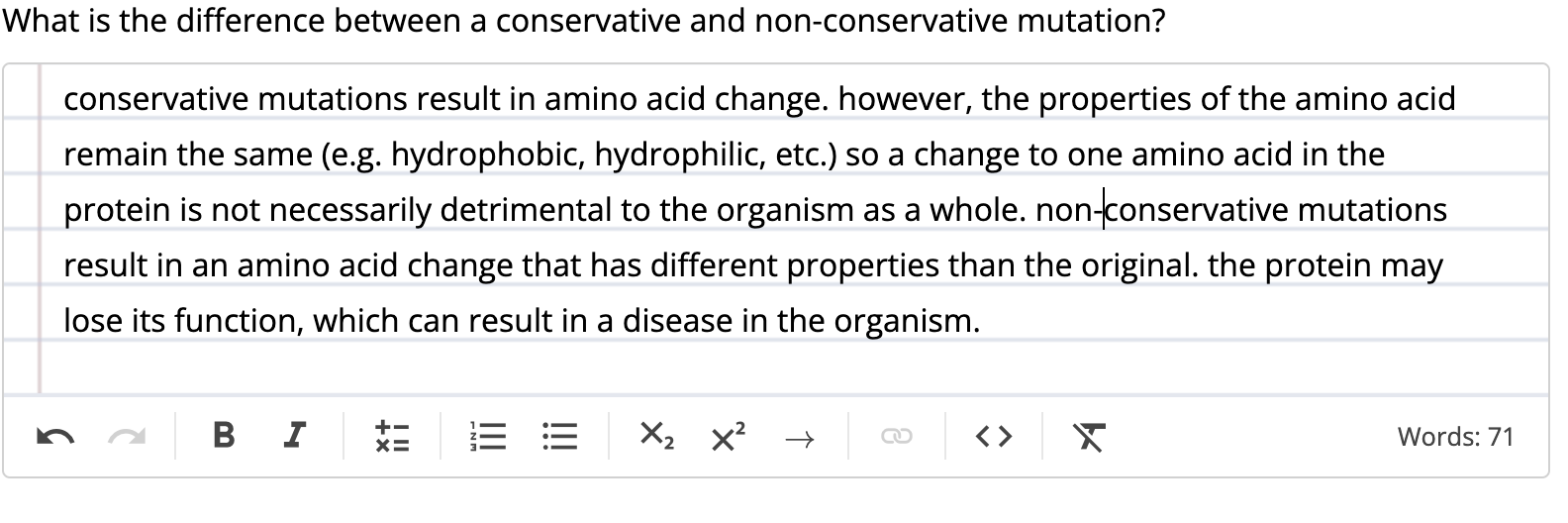
when a point mutation leads to change in amino acid, the mutation may not lead to a change in phenotype. this is called conservative mutation and involves a change from one amino acid to a different amino acid with biochemically similar properties. a non-conservative mutation results in a change to a very different amino acid, which often leads to biochemical changes.
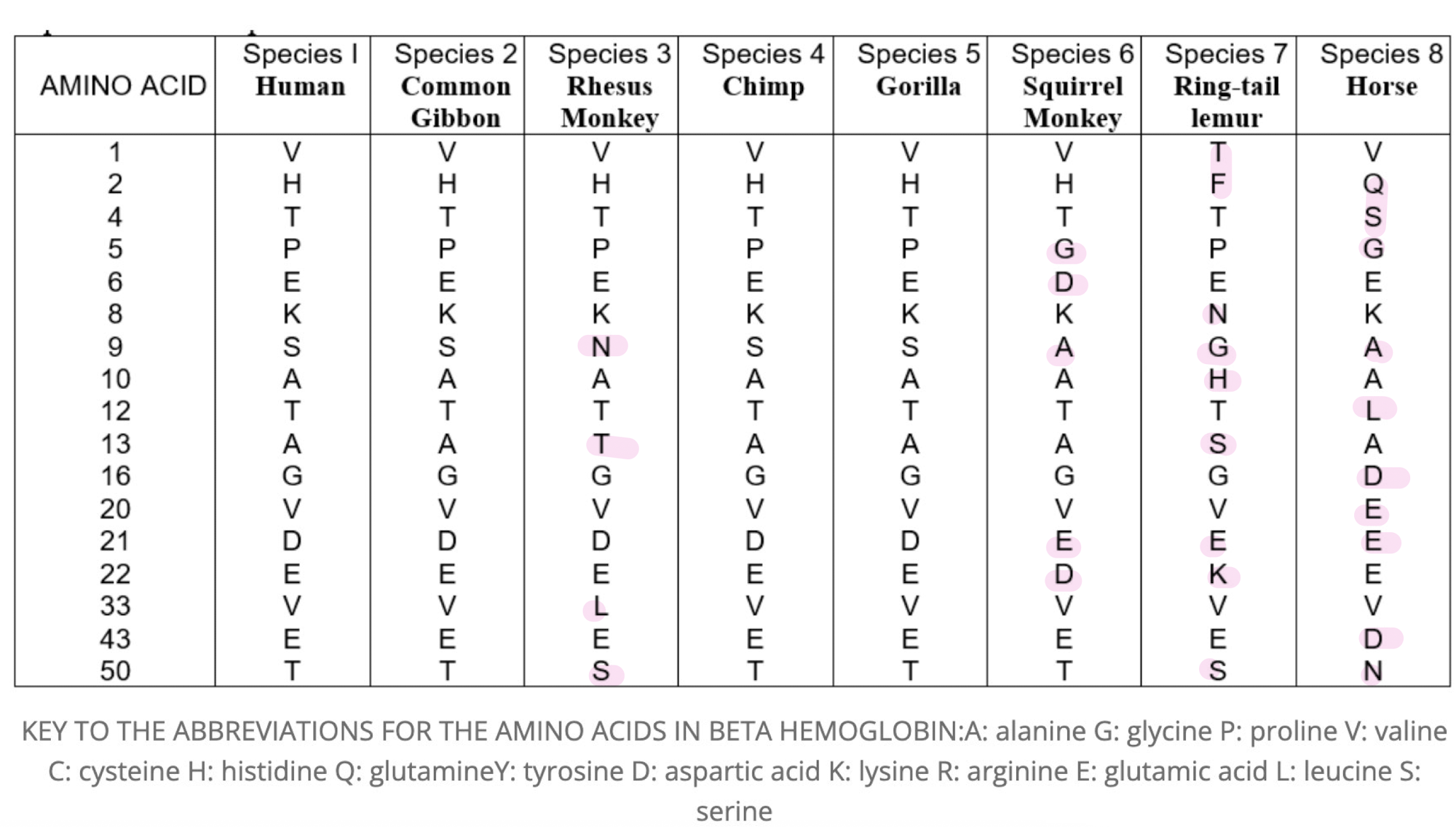
comparing common genes - the DNA of organisms can be directly compared by looking at the order of bases in comparable genes or by comparing their whole genome. closely related species will have a higher percentage of similarities in their DNA base order.
How are the genes only 76.66% similar, but the corresponding proteins are 100% similar?
conservative mutations result in the same amino acid being coded fro which results in 100% of proteins being similar.
data sheet on sac Amino Acid Comparision
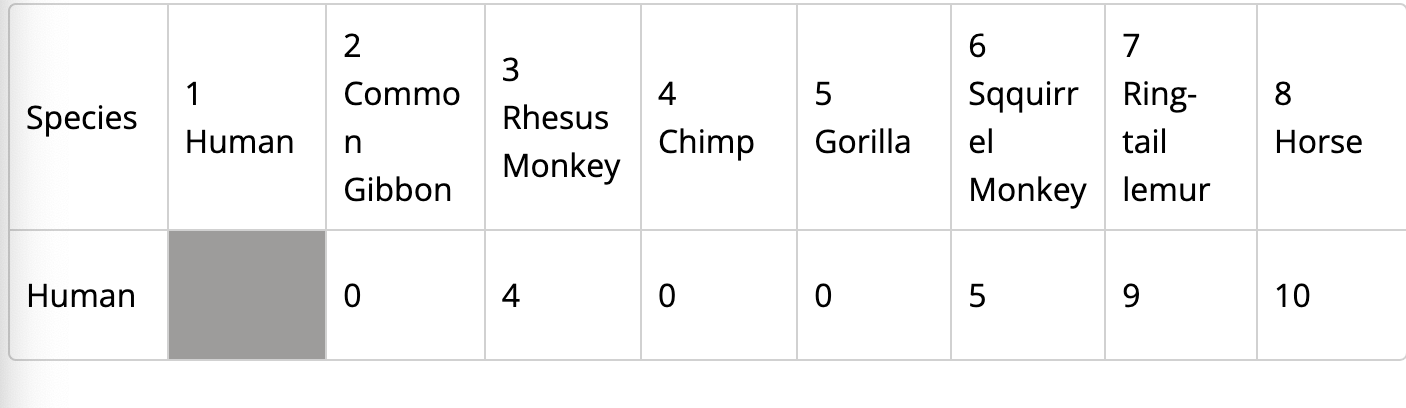
mitochondrial DNA: important tool for tracing evolutionary relationship within a species, offers several benefits over nuclear DNA when determining phylogenic pathway
maternal inheritance - mtDNA, inherited by mother only. much more direct than genetic linage
no recombination - passed from the mother. no recombination, sequence fidelity
higher mutation rate - produce reactive oxygen species, which cause sequences to mutate at a higher rate
high copy number - every cell has mitochondria, large amounts of mtDNA can be gathered fro sequencing
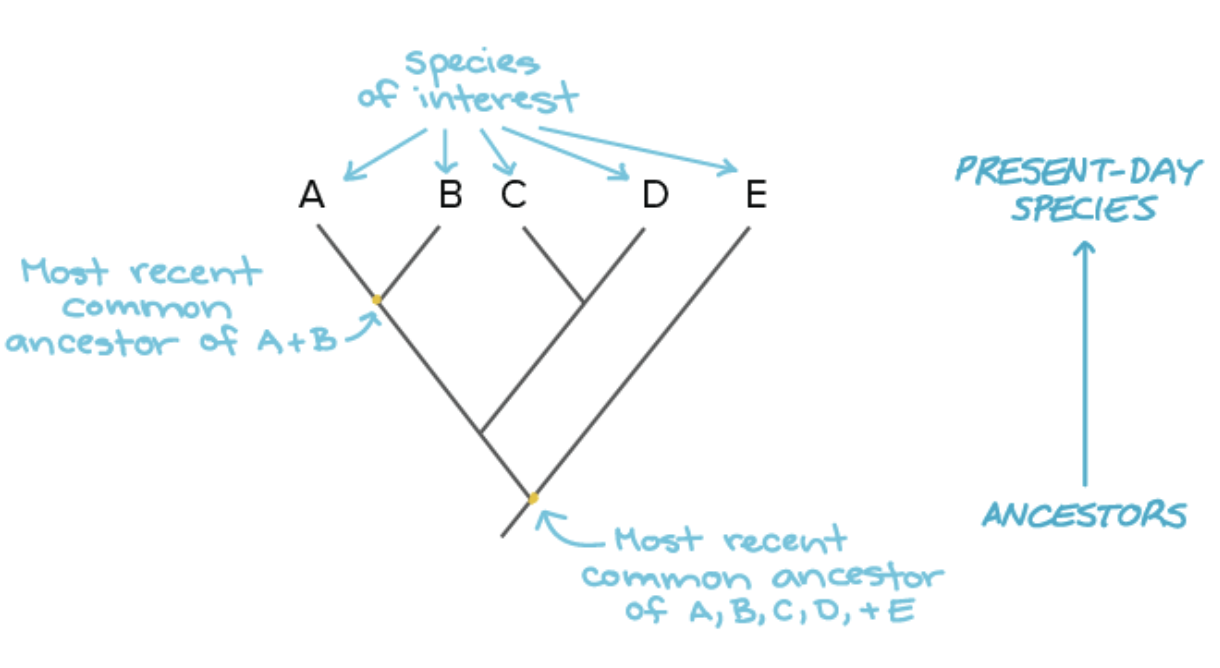
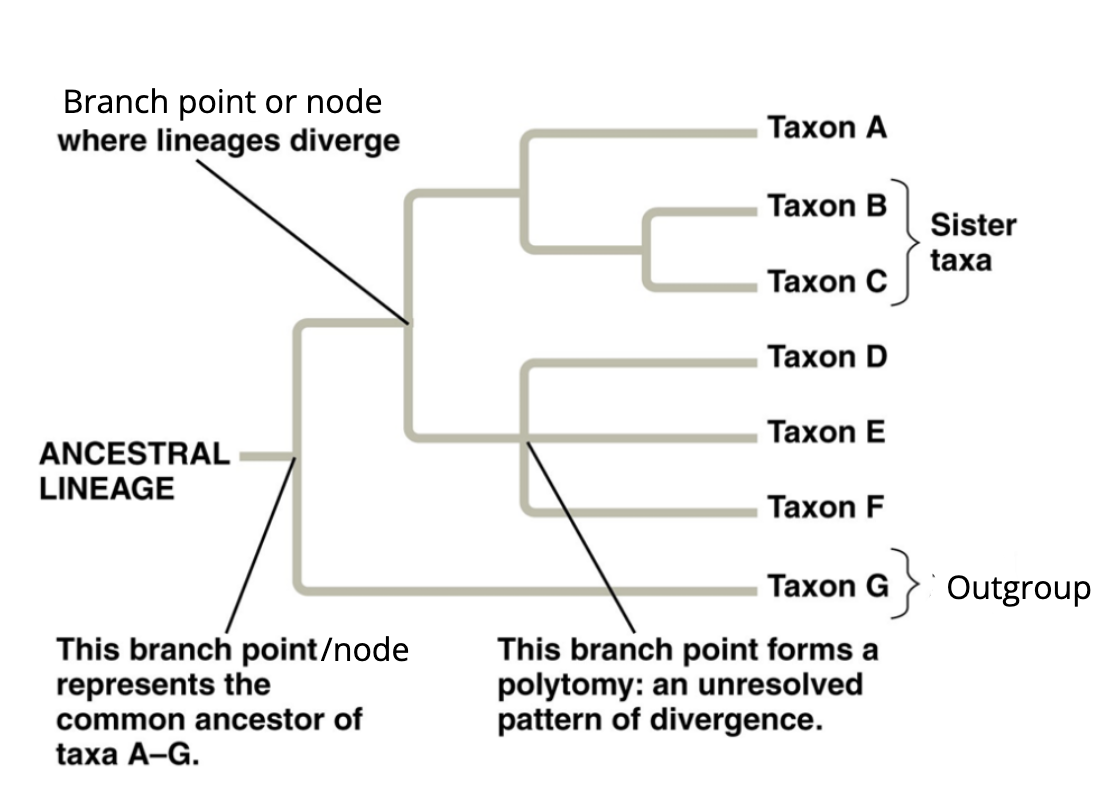
phylogenic trees
represent hypotheses about the evolutionary relationships among a group of organisms. May be built using morphological, biochemical, behavioural or molecular features of species or other groups. in building a tree, we organize species into nested groups bases on shared derived traits. the sequence of genes or proteins can be compared among species and used to build phylogenic trees. closely related species typically have few sequence differences, while less related species tend to have more.
in a phylogenetic tree, the species are shown at the tips of the trees branches and connect up in a way that represents the evolutionary history of the species. that is, how we think they evolved. each branch point lies the most recent common ancestor shared by all the species descended from that branch point.
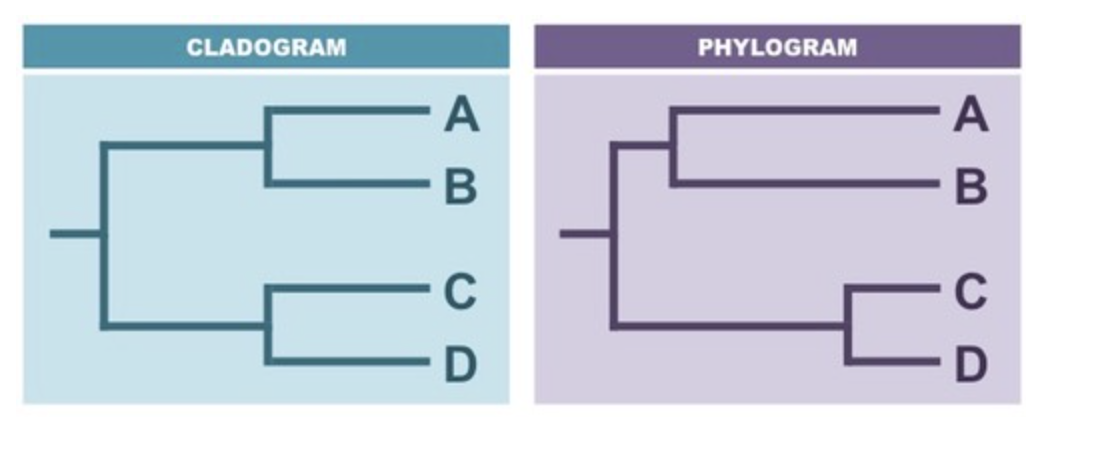
cladogram vs phylogram
cladogram give hypothetical picture of actual evolutionary history of the organism. are constructed on the basis of shared derived characteristics. the branches are of equal length as they do not represent evolutionary distance between different groups.
phylogram give an actual representation of the evolutionary history of the organisms with a time scale. the branch length are proportional to the amount of divergence
classification and characteristics
hierarchical level
class: mammals. humans are here, there are many orders within this class.
warm blooded, bear live young, females have mammary glands, specialised teeth, three middle ear bones, lower jaw, fur/fair
order: primates, humans are also here. primates include humans, great apes, gibbons, monkeys, lemurs and lorises
dextrous hands, flexible shoulder and hip joints, flattened nails that are sensitive, forward facing eyes, relatively larger regions of the brain responsible for hand-eye coordination and 3D colour vision.
superfamily: hominoids. includes humans, great apes and gibbons. note that monkeys, lemurs and lorises are not hominoids.
no tail, molars with five cusps, broad and flattened rib cage
family: hominidae. this family includes humans and great apes. gibbons are not hominids
partially or fully erect bipedal posture allowing hands to manipulate food, care for young, or use tools
subfamily: hominins. subfamily includes only humans, both surrent species and ancestral species that were bipedal (sahelanthropus, paranthropus, ardipthecus, australopithecus, homo)
bipedal - separates humans from other great apes
genus: homo modern and ancestral humans dating from appox 2 million years ago today.
species: homo sapiens
modern humans
Interpreting the human fossil records

interbreeding with Neanderthals and Denisovans: Homo Sapiens interbred with Neanderthals. How do we know? DNA analysis shows that some fossils shows a mix of both species. further testing of other fossils have shown that Homo Sapiens also mated with Homo Denisovans. So Neanderthals left Africa 100,00 years before Denisovan, because africans have no Neanderthal DNA in them.
Is Homo Denisova DNA still around today? In modern people in Philippines, Indonesia, and aboriginal Australians. Still have (4.6%) denisovan DNA in their genome today (HLA-A). This DNA was passed from mother to child, so the mother was the one to breed with the Denisovans and Neanderthals.
Megafauna and Aboriginal migration into Australia

Megafauna are large animals that roamed the Earth during the Pleistocene, 2.5 million to 11,700 years ago.
here is considerable DNA evidence (mtDNA and whole genomes) to support the hypothesis that early hominins migrated out of Africa, commencing approximately 150,000 years ago, to populate the rest of the world. DNA evidence also supports that modern hominins migrated somewhat later into Europe and the Middle East and, for a period of time, interbred with Neanderthals.
DNA evidence also suggests that modern hominins reached the Australian continent approximately 55,000 years ago from South-East Asia (although earlier dates are suggested from the dating of cultural artefacts), with distinct groups, from a single initial migration, spreading rapidly down the western and eastern coasts to occupy particular geographical areas.
The extinction of the Australian megafauna approximately 42,000 years ago is also evidence for the rapid migration of modern hominins across the continent. Evidence from DNA and cultural artefacts varies as to the amount of migration and gene flow between specific populations, however, there is evidence for prolonged connection over time of specific populations in specific areas
Our species evolved in Africa over 200,000 years ago. The Genographic Project has found that people spread out of Africa in at least two migratory waves.
The first wave travelled from eastern Africa into the area of the east coast of the Mediterranean known as the Levant about 100,000 years ago. Scientists know four species existed by 100 000 years ago: H. neanderthalensis in Europe, H. denisova in Russia, H. floresiensis in Indonesia and H. sapiens in Africa and the Middle East.
The later second wave moved from Africa into the Arabian Peninsula and continued eastward following the coast of South Asia about 70,000 years ago. This southern wave kept rolling along reaching Southeast Asia, where one branch of people migrated to Australia and New Guinea, while other branches moved along the coast of east Asia. A branch of this second wave migration moved north into the central Asia and spread west into Europe and east into Siberia about 40,000 years ago. Eventually humans made their way to the American continent about 15,000 – 20,000 years ago.
***** all numbers are ever-changing because of continuous updating and research.