Baylor BIO 2306 Miles- Unit 3
Chapter 11 - Chromosome Structure and Organelle DNA
Know the primary, secondary and tertiary levels of DNA structure
Define supercoiling and know how topoisomerases assist with it
Describe bacterial chromosomes
Define chromatin, heterochromatin, and euchromatin and differentiate between the last two (table 11.1 on page 315 is especially helpful for this)
Define histones and know the 5 types (as they relate to the nucleosome, as well as their function)
Why are they positively charged?
Describe the makeup of the nucleosome
Be able to compare and contrast bacterial and eukaryotic chromosomes
Understand basic centromere and telomere structure
Differentiate between unique-sequence DNA, moderately repetitive DNA, and highly repetitive DNA
Know the endosymbiotic theory
Know how mitochondria and chloroplasts are usually inherited, why they are inherited this way, and what that means for inheritance of those traits
Define heteroplasmy and replicative segregation (see figure 11.14 for help)
Know the characteristics of mitochondrial DNA
DNA/Chromosome Structure
Primary Structure of DNA - The nucleotide sequence
Secondary Structure of DNA - Double helix formed by hydrogen bonds
Tertiary Structure - More complex associations like supercoiling and nucleosomes
Supercoiling - a tertiary structure of DNA where it is wound tightly, dependent on topoisomerases, < (base pairs/10) rotations
Positive Supercoiling - overrotated DNA
Negative Supercoiling - underrotated, opened DNA
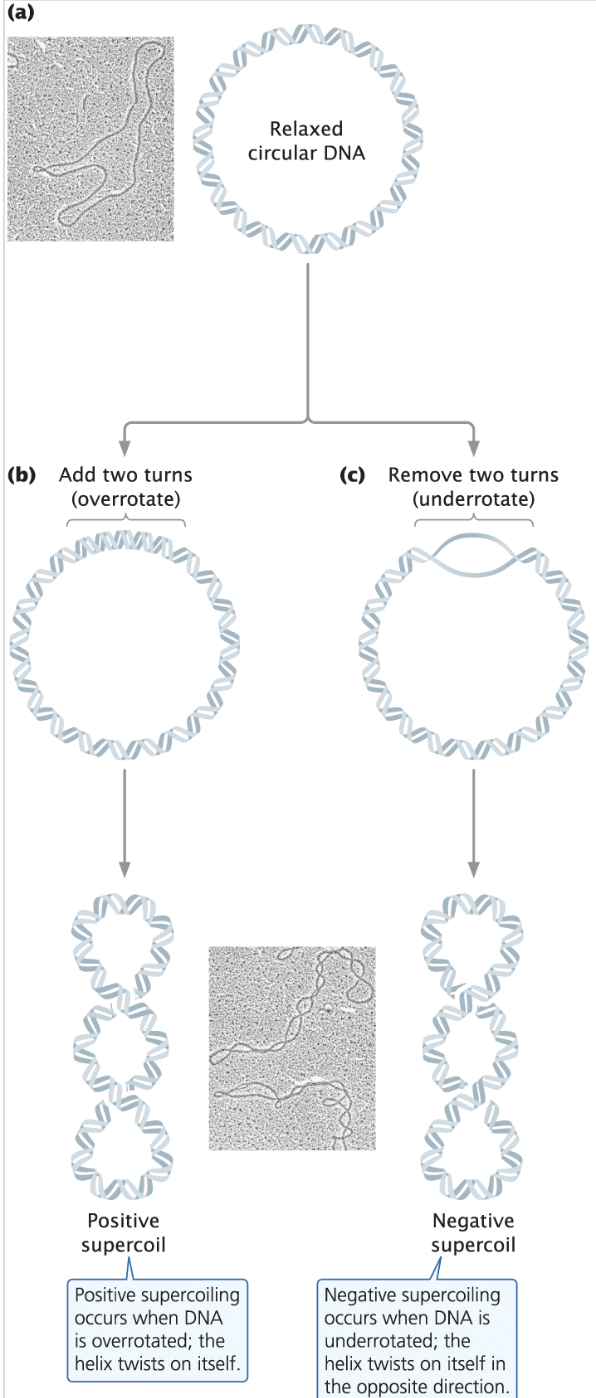
Nucleoid - a clump of DNA that works as a “nucleus” for prokaryotes
Chromatin - a complex of DNA and protein
Euchromatin - condenses and de-condenses
Heterochromatin - remains highly condense throughout the entire cell cycle
DNAse i - an enzyme that digests DNA, which reveals how chromatin structure is changed to be more exposed (to this enzyme) during transcription. Can digest opened, looser euchromatin, but not tightly wound heterochromatin
Epigenetic Changes - Alterations in chromatin structure that cause different phenotypes in the same genotype
Nonhistone chromosome proteins - Other proteins in DNA that aren’t histones
Nucleosome - the fundamental repeating unit of chromatin, consisting of a segment of DNA wound around a core of eight histone proteins.
Histones: Positively charged, H2A, H2B, H3, H4, and H1 wraps it together
Histones are positively charged because DNA is negatively charged, due to the phosphate group

Linker DNA - DNA that separates nucleosomes
Chromatin Loops - Loops of chromatin formed by loop extrusion using structural mantiance of chromosome complexes
Topologically associating Domains (TADs) - Large loops of spatially interacting chromatin that form during interphase
Polytene Chromosomes - Large chromosomes that occur when replicated DNA isn’t divided, leading to repeating, identical chromosomes put together.
Chromosome Puffs - Localized swelling of a chromosome where the DNA is opened for transcription
Centromere Structure:
Centromeres are the attachment site for the kinetocore and spindle microtubles. They are made of many base pairs of heterochromatin. They use a variant histone protein called CENP-A.
Telomere Structure:
Telomeres are the end of chromosome that are a repeating noncoding base pairs that prevent coding DNA from being degraded during replication. A multiprotein complex called shelterin binds to telomeres to prevent them from being “repaired” like a double-stranded break would be
DNA Sequences
Unique-sequence DNA - DNA present once/only a few times in the genome
Moderately repetitive DNA - long sequences that are repeated thousands of times in the genome
Tandem repeats - repeats clustered together in a chromosome
Interspersed repeats - repeats spread throughout the DNA
Short Interspersed Elements (SINEs)
Long Interspersed Elements (LINEs)
Highly repetitive DNA - short sequences that are repeated countless times in the genome.
The D loop - a region of the mtDNA that contains sites where replication and transcription of the mtDNA is initiated.
C-value - the amount DNA an organism has
C-value paradox - C-value does not equal complexity
Gene family - A group of similar/related genes; similar but not identical DNA sequences
Mitochondria & Chloroplasts
Mitochondrial Replacement Therapy (MRT) - Allowing a mother w/ mitochondrial disease to have a healthy baby using donor cytoplasm.
Endosymbiotic Theory - The theory that mitochondria and chloroplasts originated from free-living prokaryotic cells that were engulfed by a host cell and subsequently established a symbiotic relationship
Heteroplasmy - the variation of mDNA and cpDNA between multiple mitochondria/chloroplast within a cell
Homoplasmy - When 100% of mitochondria have the same DNA sequence.
Replicative Segregation - During replication, organelles segregate randomly into progeny, determining the ratio of mutant organelles to non-mutant organelles.
Denaturation - degradation of protein or DNA due to pH or temperature; split at the complementary pairs
Renaturation - Re-formation of DNA once temperature returns to normal
Hybridization - When two different DNA strands from different sources renature together due to being complementary
Chapter 12 - Replication and Recombination
Section 12.1 – None
Section 12.2
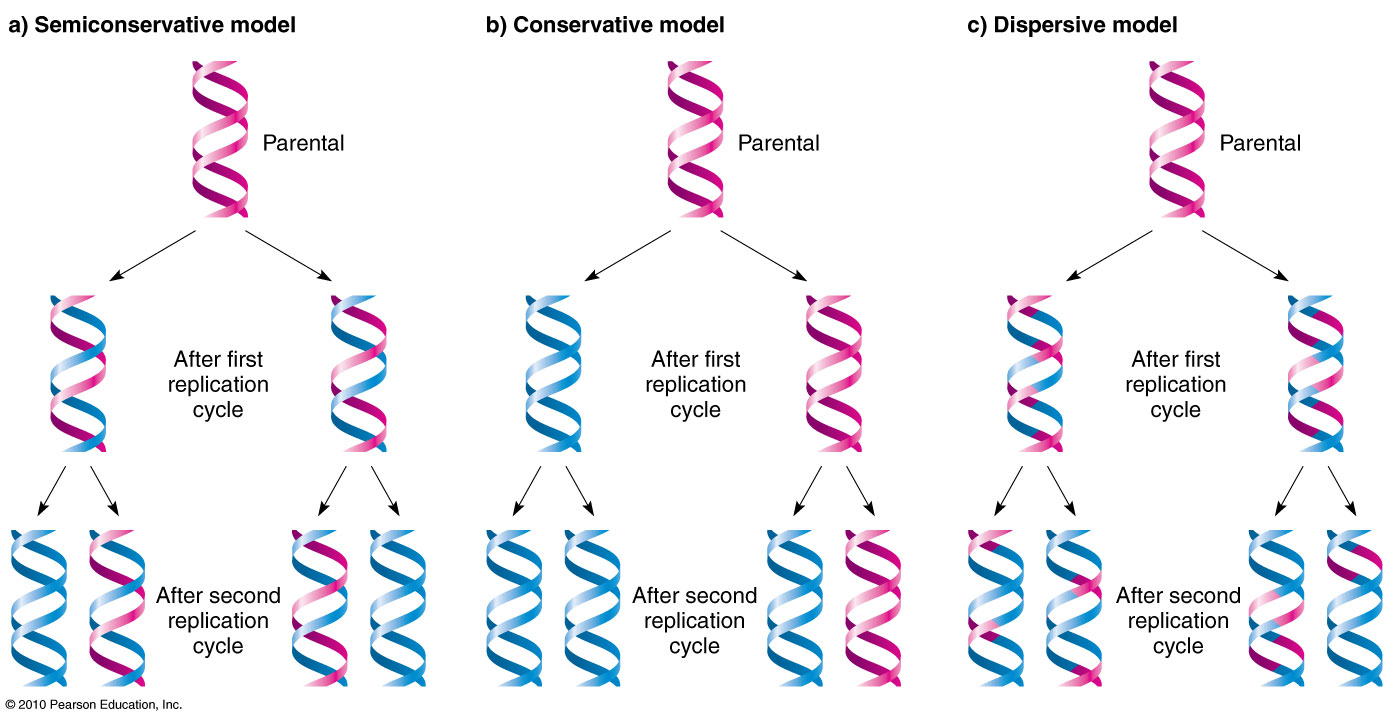
Describe the three possible models of DNA replication (conservative, semi-conservative, discursive)
Be able to predict the expected results if DNA were replicated in each of the above methods
Describe the Meselson and Stahl Experiment and which of the three modes of replications their results indicated was correct (Heavy original strands, light new strands, separated w/ equilibrium density centrifugation. semi-conservative)
How would their results have been different if DNA were replicated in one of the other ways? (conservative - no middle densities, dispersive - all the same density)
Describe the three types of semiconservative replication (Table 12.2 is a good supplementary resource) (theta (bidirectional), linear (bidirectional), rolling circle)
Know the three requirements of replication (Proteins (ex: DNA Polymerase, free nucleotides, a single stranded DNA template)
What are the raw materials (substrates) in DNA? How are they incorporated? (nucleotides: dGTP, dCTP, dATP, dTTP, 5’ phosphate groups provide energy to bind w/ 3’ hydroxyl group using DNA polymerase)
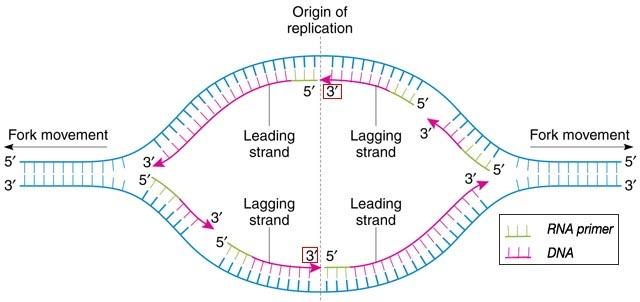
Know the direction of DNA replication (bidirectional, 5’ to 3’)
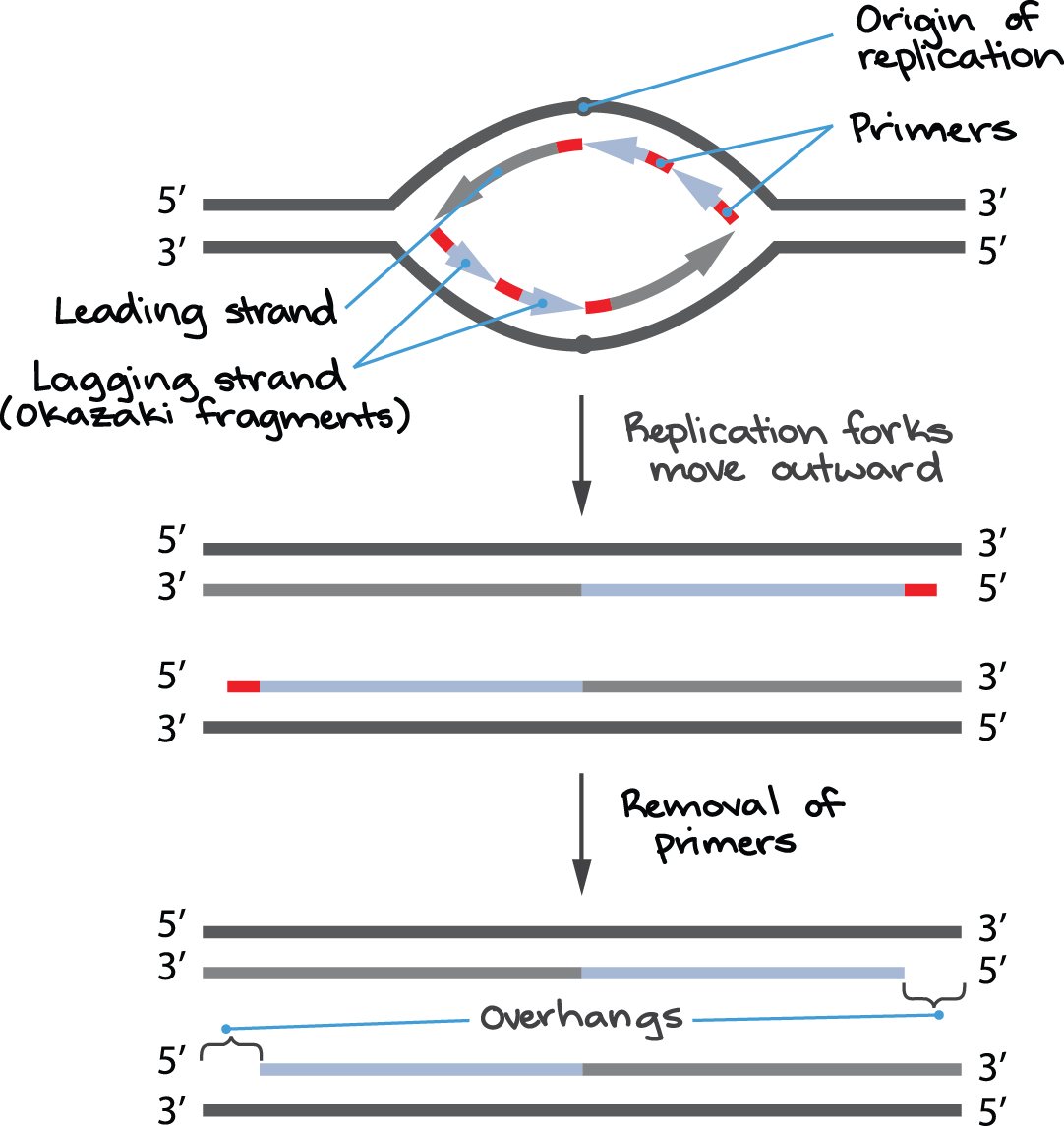
Understand and know the difference between continuous and discontinuous replication (leading and lagging strange, okazaki fragments, etc.) (See figure 12.9 for a summary)
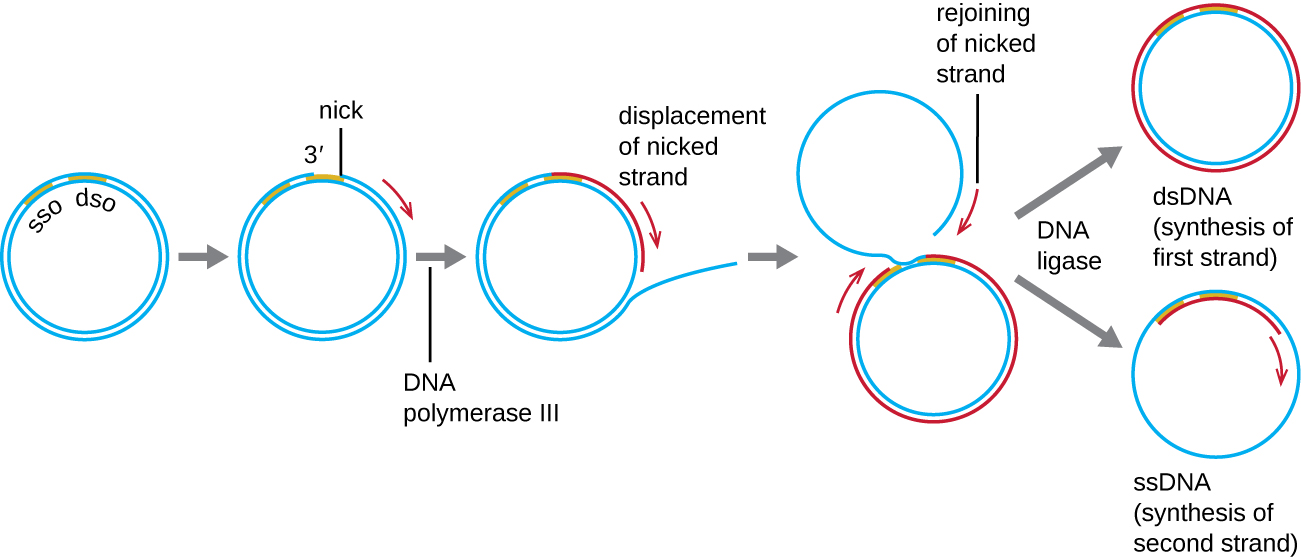
Explain (in detail!) the process of DNA replication in bacteria (this includes requirements, what happens in each stage, the key proteins and enzymes involved and their purposes, etc)
See Table 12.3 for help with the different DNA polymerases found in E. Coli
See Table 12.4 for an overview of the components required for replication in bacterial cells
Know the processes that lead to the accuracy of DNA replication
Know how eukaryotic replication differs from bacterial replication (origins of replications, DNA polymerases, process for replication the ends of the linear chromosomes, etc.)
Understand the end-replication problem and how telomerase help correct this
What types of problems could a lack or overabundance of telomerase in a cell cause?
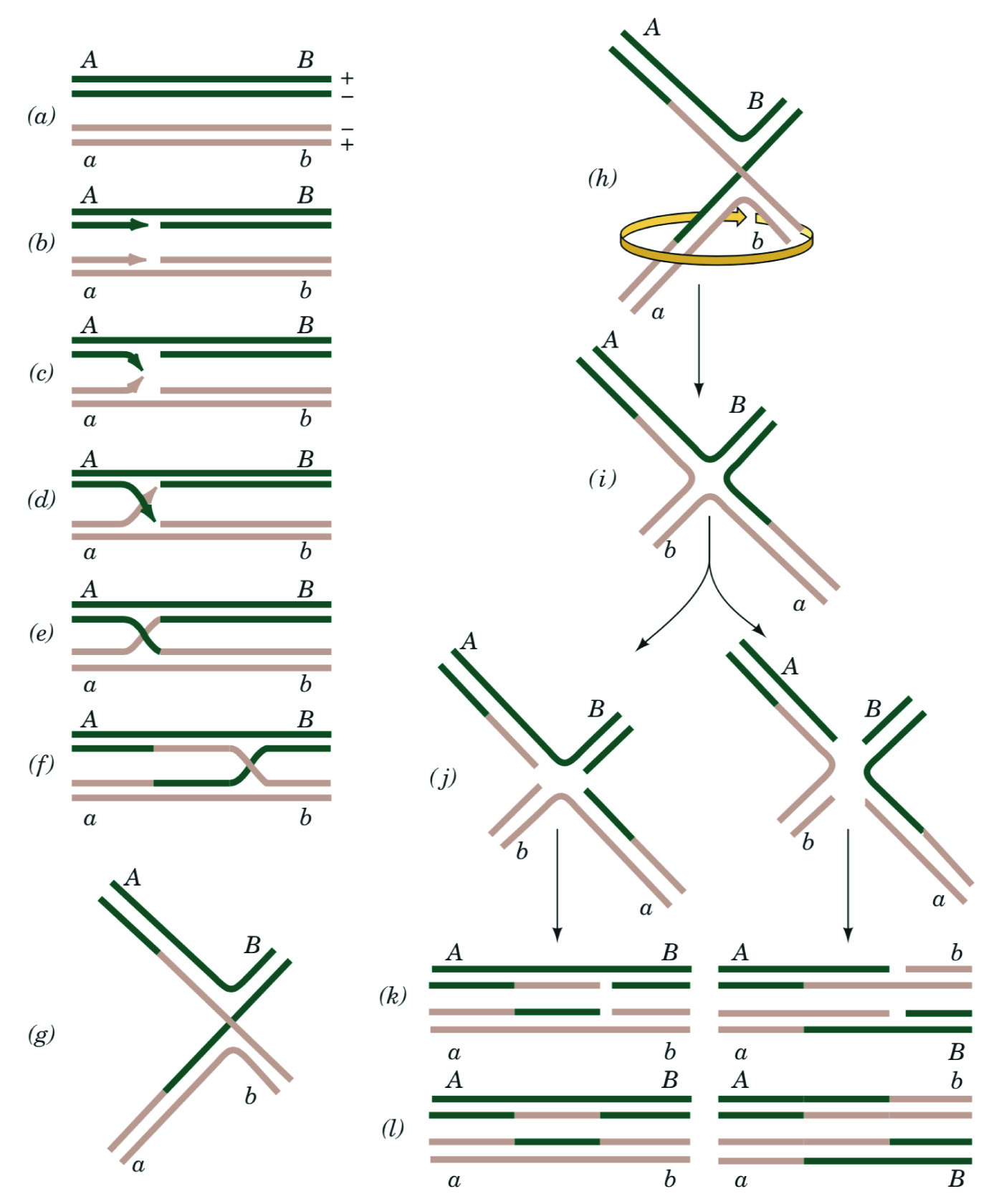
Define homologous recombination
Understand the Holliday model of homologous recombination
Problems: Comprehension Questions: 1-16
Application Questions: 19-21, 24-29, 33
Meselson and Stahl and The Semiconservative, Dispersive, and Conservative Models
Meselson and Stahl made DNA with only heavy nitrogen isotopes (15N) and allowed them to replicate with only light isotopes available for building DNA. After replication, they used Equilibrium density centrifugation to centrifuge DNA to separate the lighter and heavier DNA isotopes.
They found there was a “middle band” of medium-density DNA in addition to the light band, made partially of heavy isotopes and light isotopes, showing the semiconservative model was correct.
If the conservative model was correct, there would’ve been just a light band and heavy band.
If the disruptive model was correct, then the 3rd generation would’ve had only 1 band, as all DNA would have an equal % of the original heavy nitrogen isotopes.
Models of Replication
Important DNA structures
Replicon - The segment of DNA that undergoes replication
Origin of Replication - A segment of DNA where replication begin
Autonomously replicating sequences (ARSs) - small plasmids with origins of replication
Replication Licensing Factors - Proteins that assist in initiating replication at the origin of replication.
Replication Bubble - The opening of DNA where replication occurs
Replication Fork - the point where two DNA strands separate
DNA Nucleotides
Adenine - dATP
Thymine - dTTP
Cytosine - dCTP
Guanine - dGTP
RNA Nucleotides
Adenine - APT
Uracil - UTP
Cytosine - CTP
Guanine - GTP
Forms of Replication
Theta Replication - A form of replication that takes place in circular DNA, named because it looks like a theta halfway through replication.
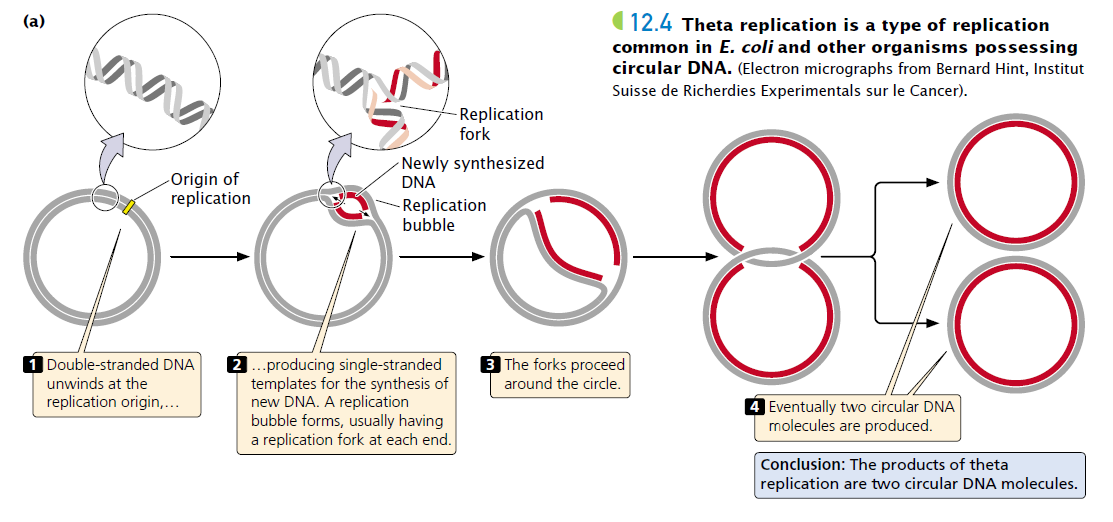
Linear Replication - standard human replication.
Bidirectional Replication - Synthesizing on both strands in different directions
Rolling-circle replication - A form of replication that takes place in circular DNA where replication “rolls” throughout the circle
DNA Replication
DNA Replication requires 3 things
A template consisting of single-stranded DNA
Raw materials (substrates) to be assembled into a new nucleotide strand
Enzymes and other proteins that “read” the template and assemble the substrates into a DNA molecule
DNA begins at the origin of replication. DNA replication at the DNA replication bubble has both continuous and discontinuous replication. continuous creates the leading strands while discontinuous leads to the lagging strands.
5’ → 3’
5’ Phosphate has a 3’ carboxyl group attached
Proteins in DNA Replication
Initiator Proteins - Start DNA replication
Single-strand-binding-proteins (SSBs) - A protein that binds to ssDNA and prevents DNA from rejoining with a complementary pair and becoming dsDNA. Used to prevent “closing” during replication.
DNA Gyrase - a topoisomerase that “untwists” the DNA, turning helix → ladder.
DNA Helicase - “unwinds/unzips” and “opens” the DNA by destroying the hydrogen bonds between nitrogenous bases
Origin-Recognition complex - Initiates replication at the origin of replication
DNA Primase - Creates RNA primers for DNA Polymerase to attach to
DNA Polymerase - Adds nucleotides 5’ to 3’ for replication
Types of DNA polymerases
Roman Numerals - Prokaryotes, Greek Letters - Eukaryotes. (flips for transcription)
DNA Polymerase III - Elongates DNA on the leading and lagging strand
3’ - 5’ exonuclease activity
DNA Polymerase II - DNA repairs, stops replication if DNA damage occurs
DNA Polymerase I - Removes and Replaces Primers (created by primase)
DNA polymerase α (alpha) - has primase activity and initiates nuclear DNA synthesis by synthesizing an RNA primer, followed by a short string of DNA nucleotides.
5’ - 3’ exonuclease activity
DNA polymerase δ (delta) - follows DNA polymerase alpha completes replication on the lagging strand.
5’ - 3’ & 3’ - 5’ exonuclease activity
DNA polymerase (gamma) - mitochondrial DNA replication and repair
5’ - 3’ & 3’ - 5’ exonuclease activity
DNA polymerase ε (epsilon) - replicates the leading strand.
5’ - 3’ & 3’ - 5’ exonuclease activity
Ligase - seals Okazaki fragments
Beta-sliding clamp - The part of the DNA polymerase that encircles DNA
Proofreading - DNA polymerase checking itself, removing mistake with exonucleases
Mismatch repair - Correcting errors in DNA
Homologous Recombination - recombination between homologous chromosomes
Heteroduplex DNA - A dsDNA segment containing strands from different sources (generally from recombination)
Holliday Junction - A special structure formed in homologous recombination
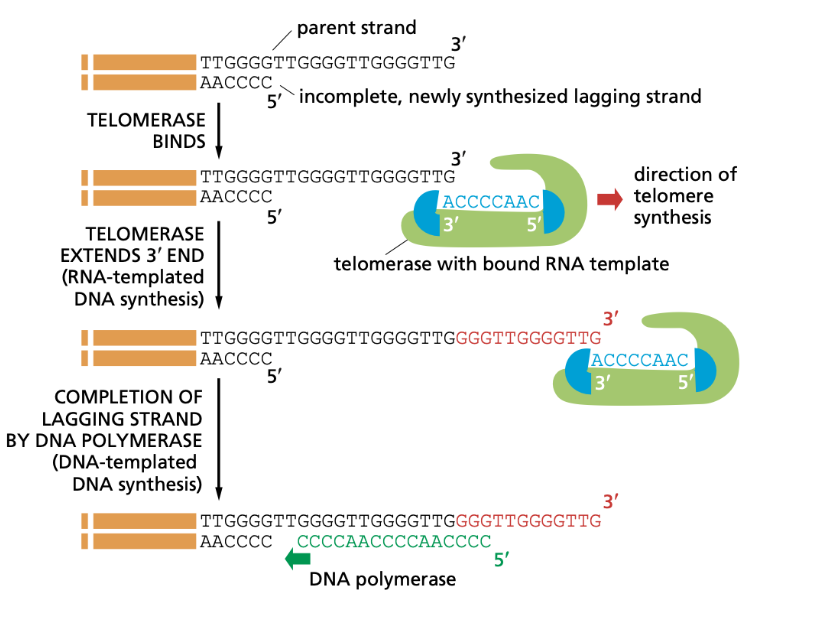
Telomeres
Telomeres - A telomere is a protective cap found at the end of a chromosome that prevents the loss of genetic information during cell division. Telomeres get smaller with each replication (see photo)
Telomerase - A protein that can extend telomerase by working as its own template and makes the telomere longer so a primer can be added further down and the overhang can be replicated. Telomerase occurs in stem cells, gametes, and cancer cells. Normal somatic cells just have their telomeres get smaller and smaller as natural aging.
G-rich 3’ Overhang - a single stranded protruding end of the telomere which allows for telomerase to extend the telomere
Chapter 13 - Transcription
Section 13.1
Explain the differences between the structure of DNA and RNA (Table 13.1 is a good resource)
List the classes of RNA and their functionality
Section 13.2
Write out the definition of transcription, the overview of its process, and the materials involved
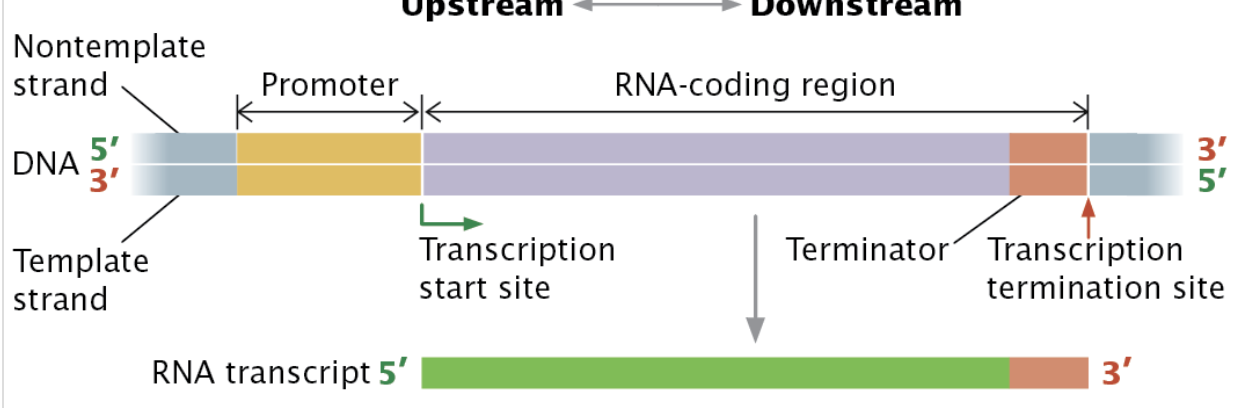
Define: transcription unit, promoter, RNA-coding region, and terminator and know how they work together; Also negative (template/non-coding) vs positive (non-template/coding) strands
Describe the bacterial transcription apparatus (figure 13.9)
Explain the differences between bacterial RNA polymerase and eukaryotic RNA polymerase
Section 13.3
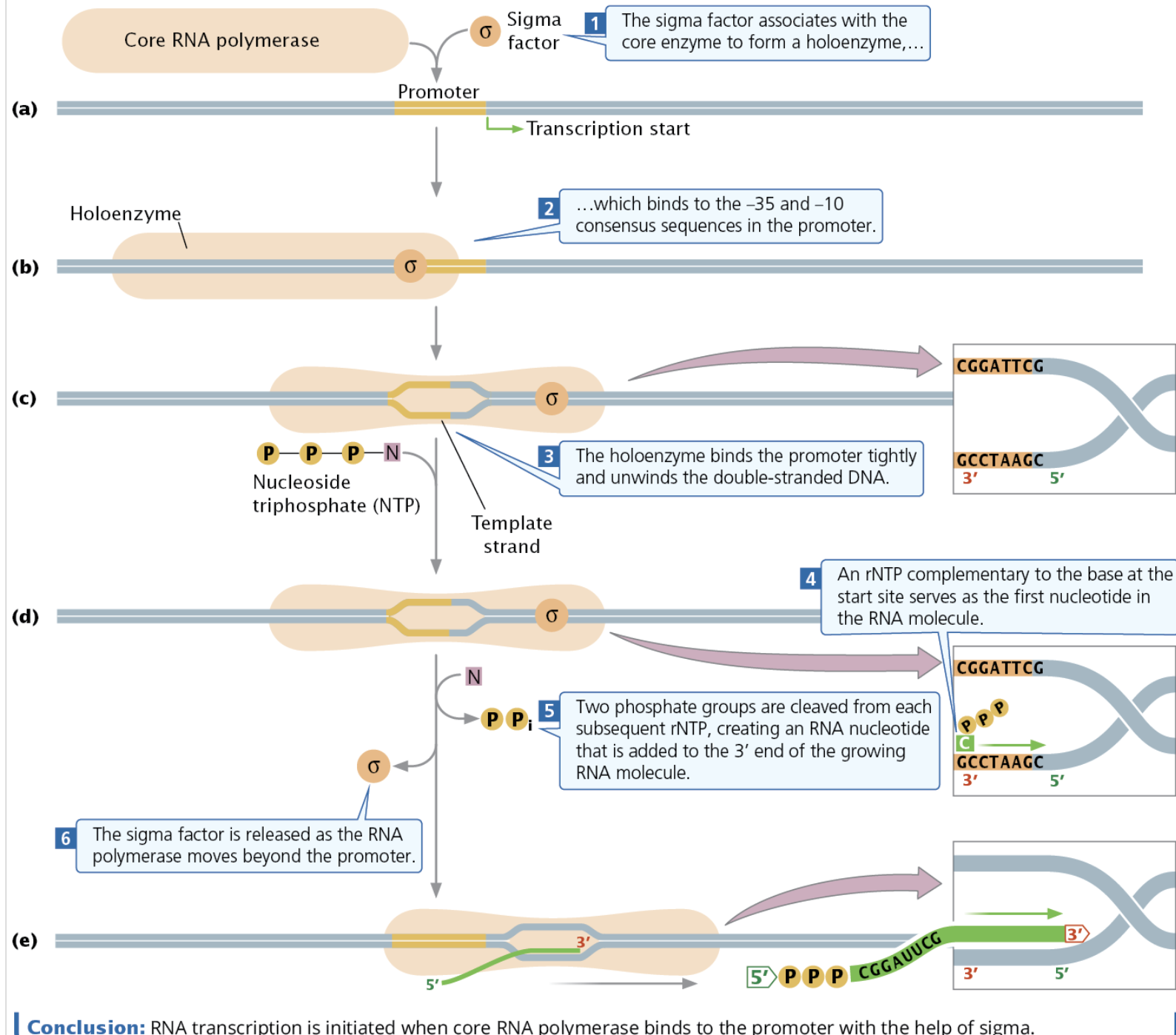
Describe the process of bacterial transcription – what proteins/factors/areas are involved in each? What are the functions of each? What enzymatic activity might they have?
Initiation
Elongation
Termination
Define consensus sequences
Section 13.4
Define and describe: general transcription factors, transcriptional activator proteins, and the basal transcription apparatus
Describe and know the differences between a core promoter and a regulatory promoter
Describe the process of eukaryotic transcription
Be able to compare and contrast bacterial and eukaryotic transcription
Section 13.5
Understand that transcription in archaea is more similar to transcription in Eukaryotes
DNA and RNA differences
DNA has deoxyribose sugar while RNA has ribose sugar
DNA has T, RNA has U
DNA has a double helix secondary structure, RNA has many types
DNA is more stable than RNA
Ribozymes - “enzymes” made of RNA
Types of RNA
rRNA (ribosomal)
mRNA (messenger)
pre-mRNA (pre-messenger RNA with intron sequences)
tRNA (transfer)
snRNA (small nuclear)
Combine with protein to create snRNPs
snRNPs (small nuclear ribonucleoproteins, “snurps”)
Process/cut pre-mRNA
snoRNAs (Small nucleolar RNAs)
Help process rRNA
microRNA (miRNA)
Help with RNA interference (RNAi)
Small interfering RNA (siRNA)
Help with RNA interference
Piwi-interacting RNAs (piRNA)
Interact with piwi proteins, similar to siRNA and miRNA
Long noncoding RNA (lncRNAs)
Its in the name
CRISPR RNAs (crRNAs)
Assist in destruction of foreign DNA
Transcription Unit - A stretch of DNA that encodes an RNA molecule that includes a promotor and terminator.
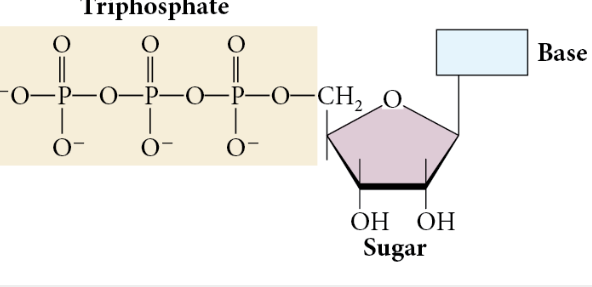
Ribonucleoside Triphosphate (rNTPs) - The molecule that synthesizes RNA made of a base, a sugar, and triphosphate.
Core Enzymes - Two copies of a subunit that make up most of bacterial RNA polymerase
Sigma factor - In Bacteria, a part of RNA polymerase that controls binding of RNA polymerase to the promoter and initiates bacterial transcription
Holoenzyme - Sigma factor + core enzyme
Types of RNA Polymerases
Eukaryotic RNA polymerases (numerical, opposite of DNA Polymerases w/ greek)
RNA Polymerase I - Transcribes rRNA
RNA Polymerase II - Transcribes pre-mRNA, snoRNAs, miRNAs
RNA Polymerase III - Transcribes tRNAs, rRNAs, mrRNAs, siRNAs
I, II, III are found in all eukaryotes, IV and V are found in plants
RNA Polymerase IV - Transcribes siRNAs and silences transposons (genes that move around a genome)
RNA Polymerase V - Transcribes siRNAs
Prokaryotes only have one RNA polymerase
Consensus Sequences
Consensus Sequence - a DNA sequence that highlights the most common or conserved nucleotide or amino acid at each position across a set of aligned homologous sequences. (Ex: comparing the sequence of the same gene in horses, humans, and cats and seeing what nucleotides most common)
-10 Consensus Sequence (Within the TATA box) - the most commonly encountered consensus sequence found in almost all bacterial promoters, found 10 bp before the coding sequence
-35 Consensus Sequence - Another common bacterial promoter sequence, found 35 nucleotides before the coding sequence
Upstream element - A consensus sequence in the promotor
Abortive Initiative - When RNA polymerases generates and releases short transcripts, failing to properly initiate transcription. It often takes several abortive initiatives for proper transcription.
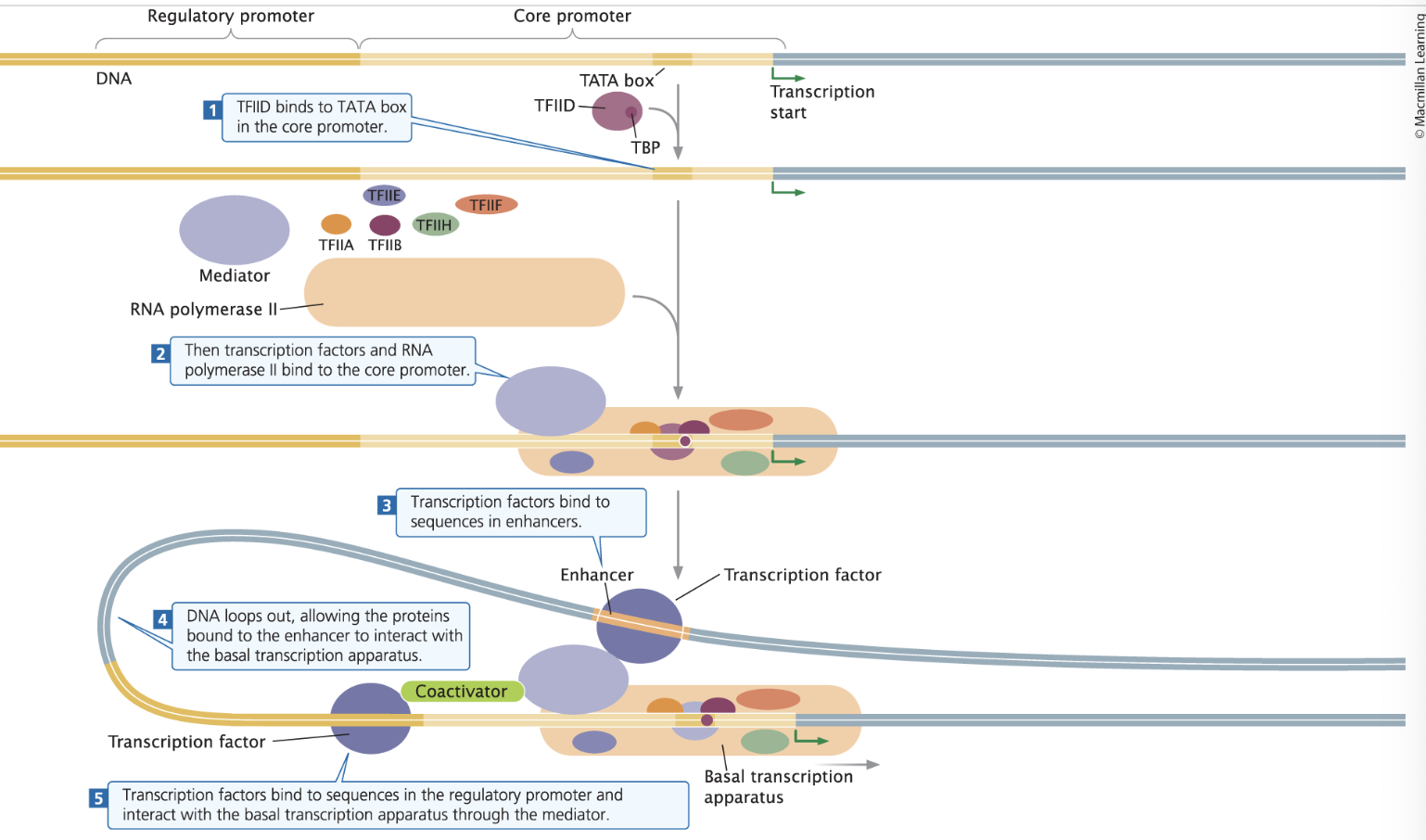
RNA Polymerase II Transcription Initiation
→ 1) Important Sequences/Areas
Regulatory Promotor - Located upstream of (before) the core promotor and impact the rate of transcription (regulate)
Core promoter - Located downstream of (after) the regulatory promotor; The site where the basal transcription apparatus binds
TATA box - a common sequence in the core promoter
Enhancers - Distant DNA sequences that can be bound to transcription factors to increase transcription
→ 2) The Proteins
Transcription Factors - Proteins that bind to DNA sequences and affect levels of transcription
General Transcription Factors - transcription factors that form the basal transcription apparatus
TATA-binding proteins (TBP) - a protein that binds to the TATA box that helps “open/unwind” the ds DNA
TBP
TFIID
Others
TFIIE
TFIIA
TFIIH
TFIIF
Basal transcription apparatus - A group of proteins that assemble near the transcription state site that can initiate minimal levels of transcription in DNA Polymerase II
Non-template Strand - A coding strand of DNA, the strand that is NOT transcribed, aka…
Positive Strand
Coding Strand
Sense Strand
Template Strand A non-coding strand of DNA the, 3’ to 5’, aka…
Antisense Strand
Negative Strand
Noncoding Strand
→ 3) The Process
The Transcription Factors TFIID (aka TF2D) binds with TBP, where they then attach to the TATA box in the core promotor.
Then, RNA Polymerase II combines with TFIIA, another TFIIB, TFIIE, TFIIH, TFIIF. The mediator helps communication between the RNA Polymerase II and the transcription factors.
A transcription factor binds to the far-away enhancer and gets attracted and bound the the RNA polymerase.
The transcription factors and RNA Polymerase II form the Basal Transcription Apparatus, which is assisted by the mediator (now called the coactivator). Transcription begins building 5’ to 3’, reading 3’ to 5’.
Elongation
Similar to replication, just going down the DNA template.
Prokaryotic Terminators
Rho-dependent terminators - terminators dependent on an ancillary protein called rho factor (p); the rho attach to the rut sequence and then unbinds the RNA polymerase from the DNA.
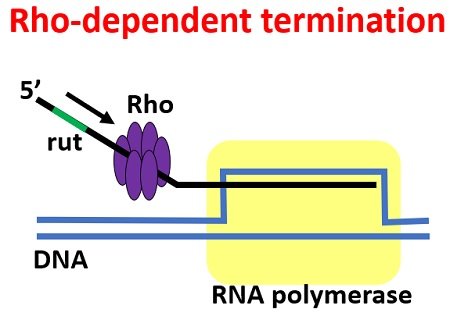
Rho-independent terminators - terminators that don’t need rho factor
Polycistronic mRNA - A group of bacterial genes transcribed into a single mRNA
Eukaryotic Termination:
RNA Polymerase I Termination: Termination requires a termination factor that binds to a sequence downstream of the termination
RNA Polymerase III Termination: Ends after transcribing a terminator sequence that produce a string of uracil molecules, sometimes needing hairpins/secondary structures
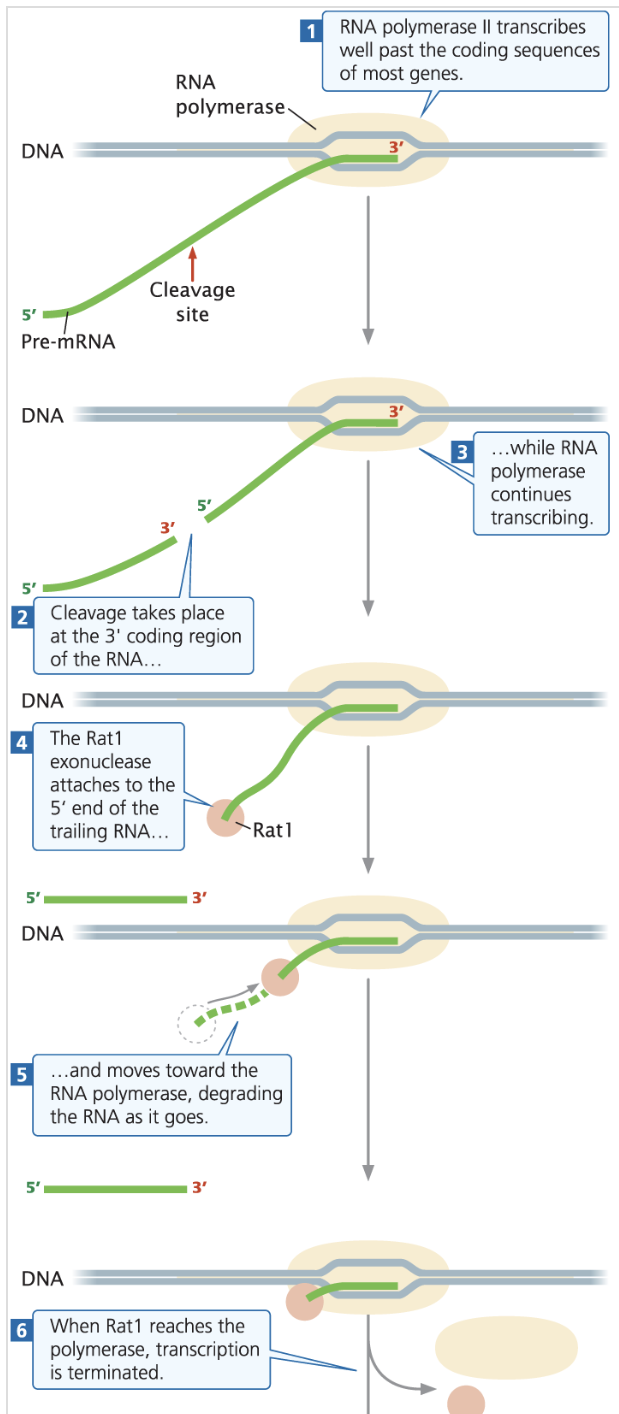
RNA Polymerase II Termination: Transcribes well past the coding sequence, more being cleaved and degraded back towards the terminator via exonuclease like RAT1 (5’→3’)
Exonucleases - enzymes that cleave nucleotides one at a time from the ends of a nucleic acid polymer, either the 5’ or 3’ end
RAT1 - a 5’→3’ exonuclease found in yeast
Chapter 14 - RNA Molecules and RNA Processing
Section 14.1
Write out the definitions and differences between introns and exons
Section 14.2
Draw the structure of a bacterial mRNA (Reference figure 14.5 if needed)
Explain the three main steps of pre-mRNA processing in eukaryotes; Include what is meant by posttranscriptional modification, in what cells they occur, and where they occur in the cell
Describe the types of eukaryotic pre-mRNA processing listed in table 14.2, the processes behind them, and their functions
Describe the process of splicing (figures 14.8, 14.9); Define spliceosome and its components
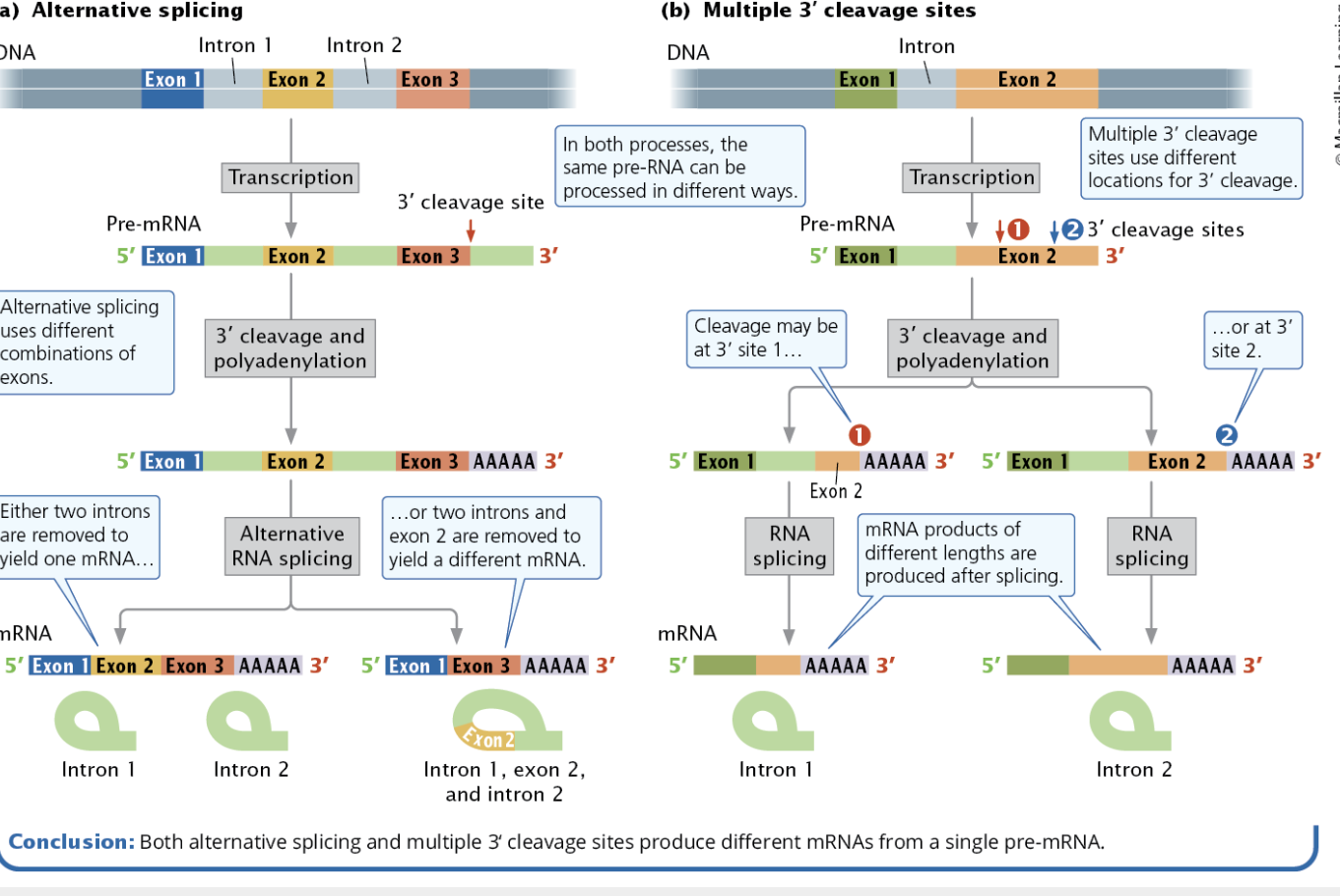
Define and describe alternative splicing (figure 14.12)
Define and describe multiple 3’ cleavage sites (figure 14.12)
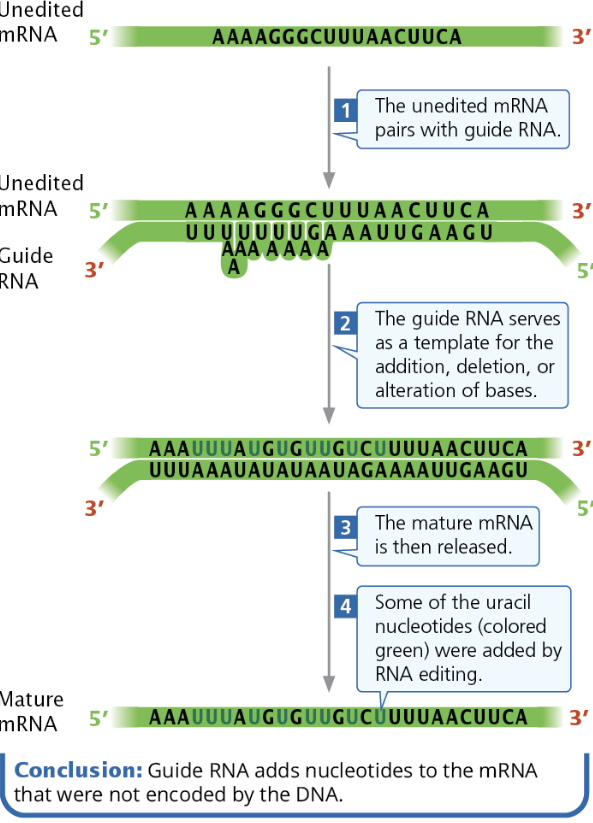
Define and describe the process of RNA editing
Section 14.3
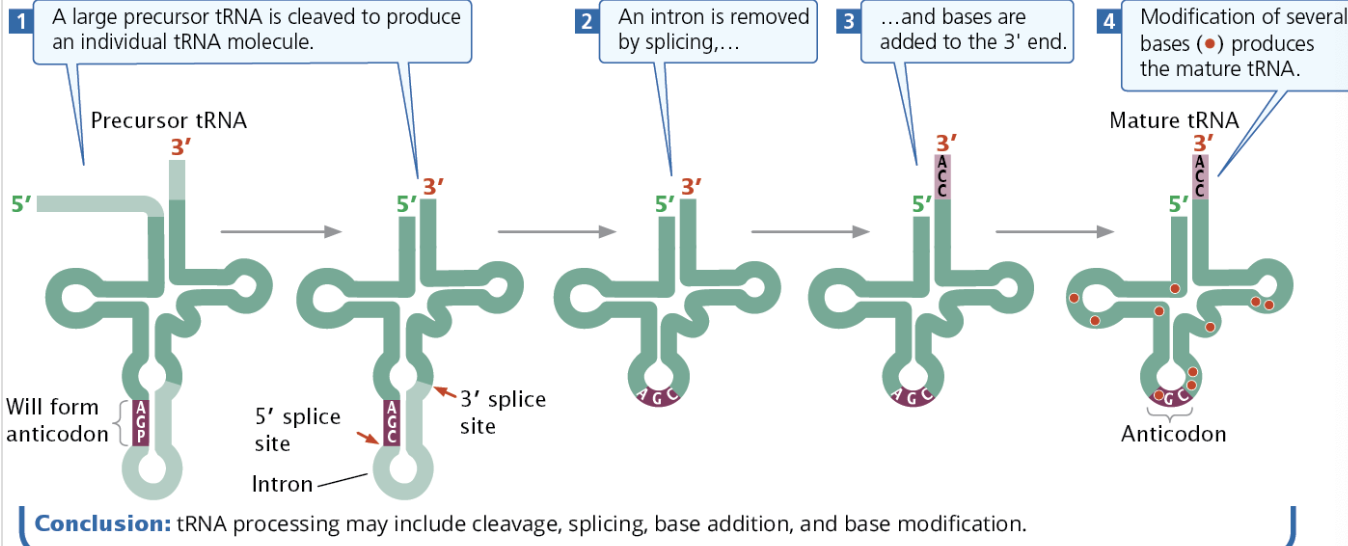
Draw and explain the function and structure of tRNA; Describe tRNA processing and what each tRNA must have (figure 14.19, 14.20)
Section 14.4
Explain the functions and general structures of rRNA
Describe the structure of a ribosome; Describe rRNA processing (figure 14.21)
Section 14.5
Describe how RNAi works (figure 14.22) and its purpose; miRNA vs siRNA form and function
Describe CRISPR RNA and its purpose
Section 14.6
None
Colinear - A term describing the direct relationship between DNA sequences and the amino acid sequence of the corresponding protein.
Introns & Exons
Exons - The coding regions of DNA
Introns - noncoding regions of DNA, also called “intervening sequences.” These sections are removed from pre-mRNA to make mRNA. Introns are common in Eukaryotes, but rare in Bacteria.
Intron Early Hypothesis - The idea that early organisms had introns and they were lost in prokaryotes
Intron Late Hypothesis - The idea that early organisms did not have introns and eukaryotes later acquired them
Group I Introns - Self-splicing introns found in bacteria, bacteriophages, and eukaryotes
Group II Introns - Self-splicing introns found in mitochondria, chloroplasts, some bacteria, and archaea.
Nuclear pre-mRNA introns - Non-self-splicing (requires a spliceosome) introns found in the eukaryotic nucleus
Transfer RNA Introns - Non-self-splicing introns found in tRNA genes of bacteria, archaea, and eukaryotes
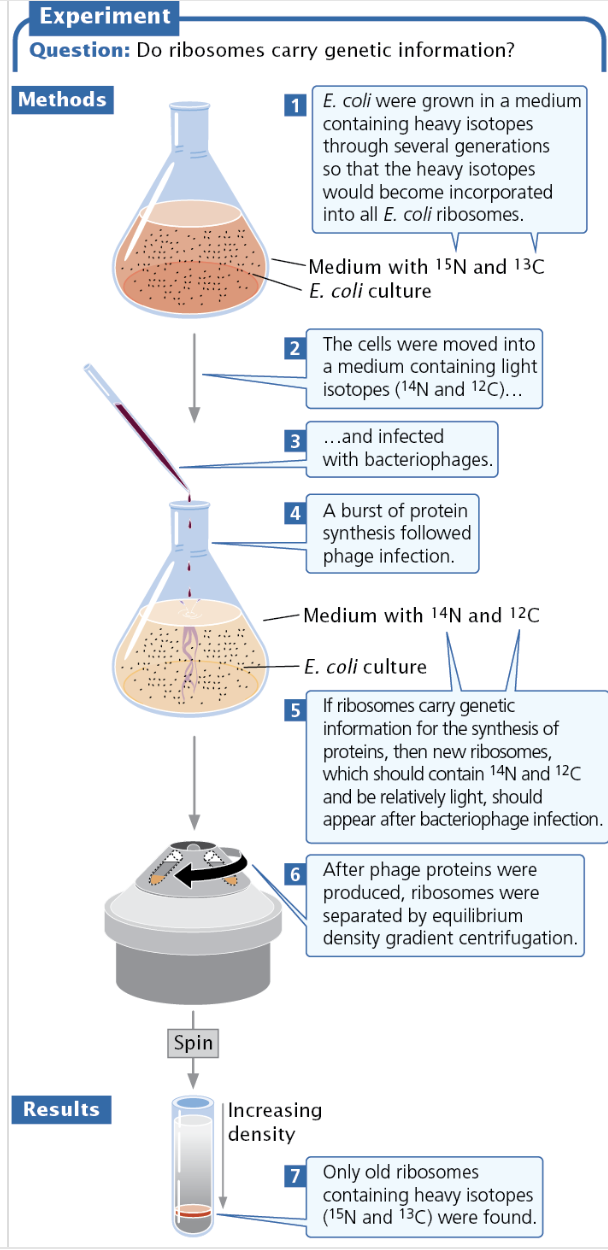
Brenner, Jacob, and Meselson Experiment
Brenner, Jacob, and Meselson wanted to see if ribosomes carried genetic information.
They added heavy nitrogen and carbon isotopes (N15 and C13) to ribosomes and moved them into a medium with only light isotopes (N14 and C12).
If Ribosomes carried DNA, then new ribosomes with the light isotopes would be found, but when they used equilibrium density gradient centrifugation they found only the old, heavy ribosomes.
The Structure of Bacterial Messenger RNA
mRNA is a template for protein synthesis, carrying genetic information from DNA to ribosomes. mRNAs have codons, which are sets of three bases/nucleotides which translate to a specific amino acid.
The 5’ untranslated region (5’ UTR) is the sequence of nucleotides at the 5’ end that first “go into” the ribosome.
In bacteria, this region is a consensus sequence called the Shine-Dalgorno sequence and serves as the ribosome-binding site on the small subunit for translation.
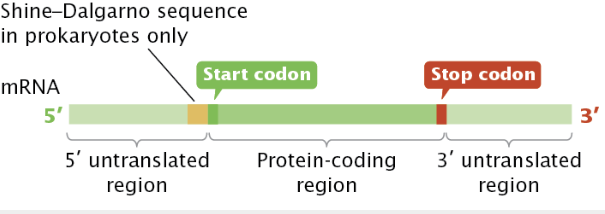
After the 5’ UTR is the protein coding region which starts with an start codon and is translated. It ends with a stop codon. Then, there is the 3’ untranslated region (3’ UTR) which impacts mRNA stability and helps regulate mRNA translation.
Pre-mRNA Processing
In bacteria, transcription and translation occur simultaneously— the 5’ end is being translated while the 3’ end is still being transcribed. There is no mRNA processing.
However, eukaryotic mRNA must travel from the nucleus to a ribosome, giving it both time to be processed and the potential to be degraded. Like transcription, pre-mRNA processing occurs in the nucleus.
→ Types of Eukaryotic processing
1) Adding 5’ Guanine Cap - increases mRN stability and helps with splicing
Only place with 5’ → 3’ linkage
2) 3’ cleavage and Poly-a tail (adenine) addition - increases mRNA stability, helps with transport, helps with ribosome binding
Length is determined by how long mRNA needs to survive
3) RNA splicing - removes introns and allows for multiple proteins to be produced through
Occurs in the nucleus
Splicing is done by a spliceosome
+ RNA editing - alters sequence of mRNA
+ Internal RNA modifications - Influences splicing, degradation, and translation
Most commonly adding a methyl group
→ RNA Splicing
Splicing requires an intron to have a 5’ splice site (first cut), a 3’ splice cut, and and a branch point made of an adenine nucleotide.
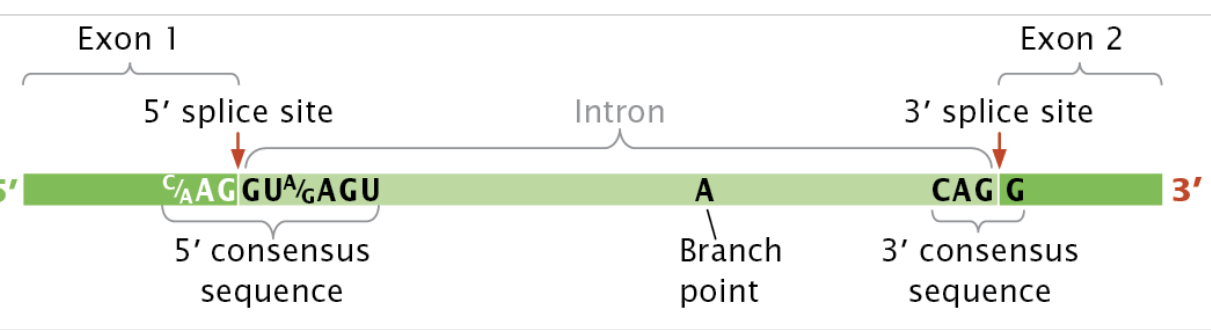
Splicing at the 5’ site leads to the 5; site attaching to the branch site to form a lariat within the intron
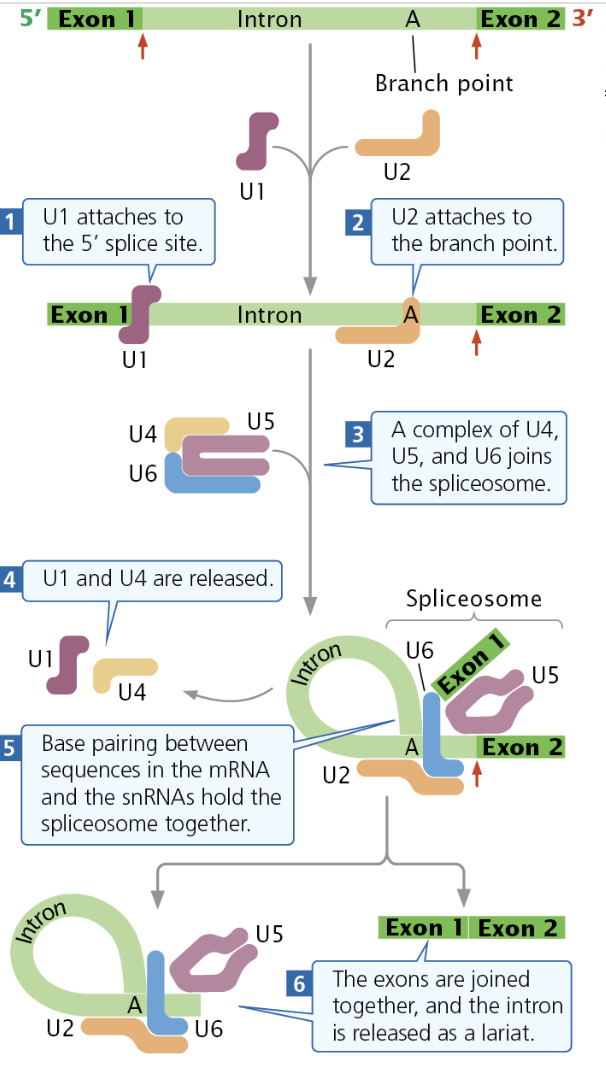
Lariat Formation/Splicing Process
The Spliceosome is made of several snRNPs and other components
First, snRNP U1 attaches to the 5’ splice site, while snRNP U2 attaches to the branch point.
The U4, U5, and U6 complex, attach to the intron, forcing the U1 to loop over and the 5’ end will be cut
Then, U1 and U4 detach, causing U2, U5, and U6 to base pair together, creating the spliceosome
The spliceosome cuts the 3’
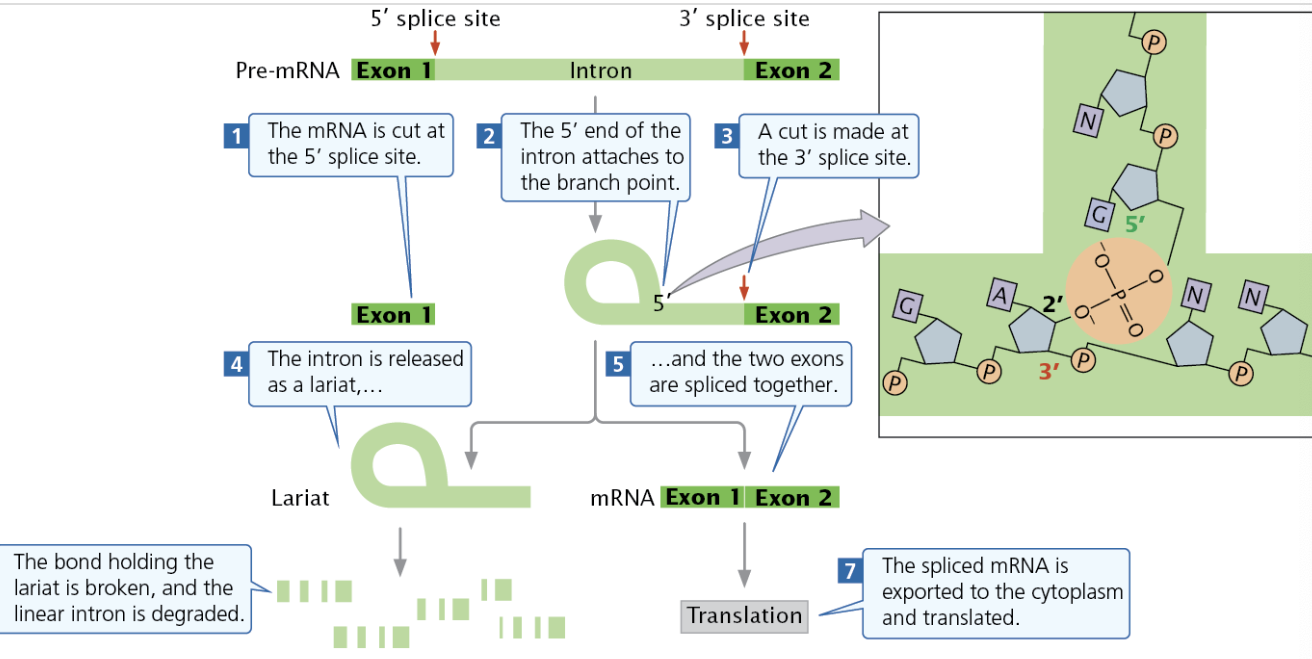
→ Minor Splicing
Some small introns in multicellular eukaryotic pre-mRNA is spliced by minor splicing, where the spliceosome used is made of different snRNAs.
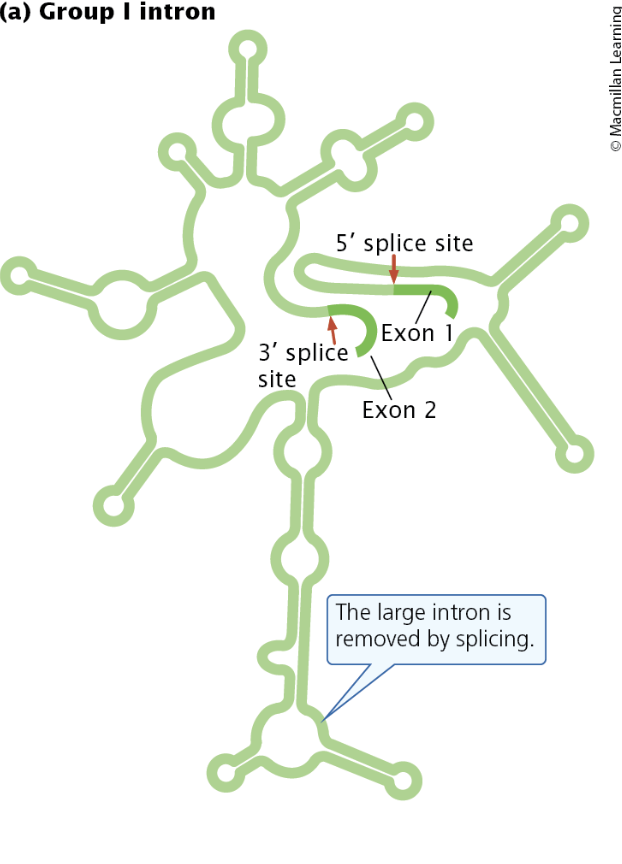
→ Self-Splicing Introns
Self-Splicing introns such as Group I and II introns create secondary structures such as hairpins which are essential for splicing.
Alternative Processing Pathways
Alternative processing pathways allow a single pre-mRNA to be processed in multiple ways to produce different proteins.
One form of this is alternative splicing where pre-mRNA can be spliced in different ways, leaving different nucleotide sequences and therefore different amino acid sequences.
Trans-splicing - Splicing together two different mRNAs
→ RNA Editing
RNA editing changes the coding sequence of mRNA after transcription. Guide RNAs (gRNAs) are partially complementary to segments of unedited mRNA and the gRNA binds to mRNA. Then, changes are made to mRNA following the gRNAs template
Structure of Transfer RNA
tRNAs have modified bases, allowing them to have more base types that occur only in tRNAs (except psuedodouridine, which occurs in rRNAs and snRNAs.) Modified bases are created by tRNA-modifying enzymes.
Standard RNA bases
Uracil (U)
Guanine (G)
Adenine (A)
Cytosine (C)
Additional Bases (from uracil)
ribothymidimine
psuedouridine
tRNA has a “cloverleaf”-like secondary structure with four major arms, the anticodon arm, the amino acid attachment site, the TΨC arm, and the DHU arm. The anticodon is paired with a codon in the ribosome.
tRNA contain introns like mRNA and need to be spliced before becoming mature tRNA.
Ribosomal Structure
Ribosomes are made of a large ribosomal subunit and a small ribosomal subunit.
rRNA is spliced apart by snoRNAs like mRNA.
RNA Interference
RNA Interference contributes significantly to gene significantly. dsRNA triggers RNAi. One way it does this causing transcription of an inverse sequence than then binds to a normal transcript which prevents it from being translated.
Dicers: chopped by dsRNA. The smaller, resulting RNA fragments are opened/unwound to make siRNAs and miRNAs.
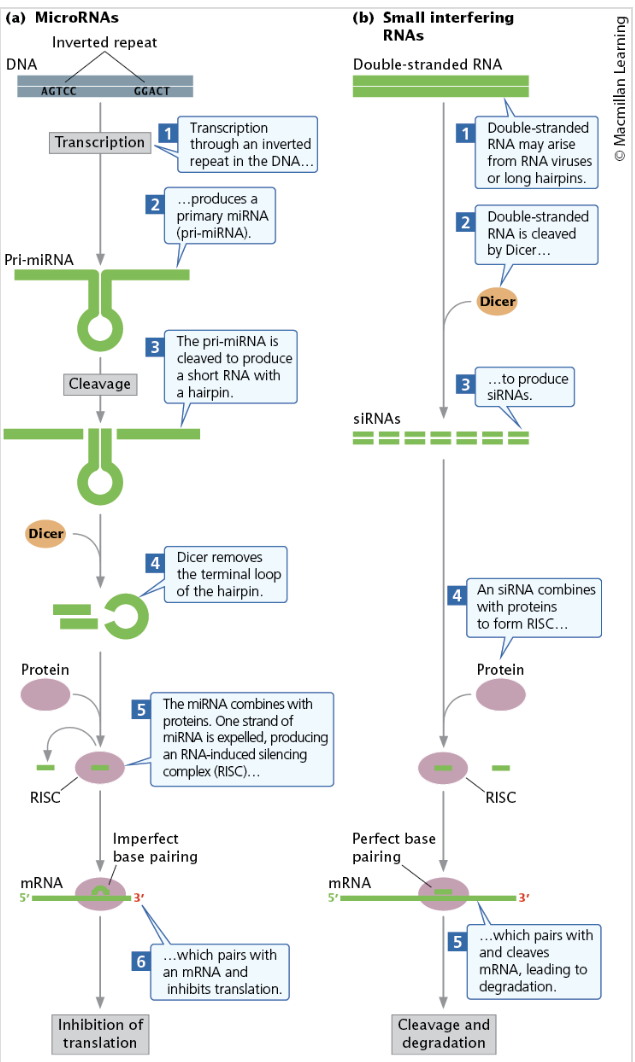
siRNA and miRNAs combine with proteins to form RNA-induced silencing complex.
While miRNA come from distinct genes, siRNA come from mRNA, transposon (movable genes), or viruses. miRNA degrade mRNA and inhibit translation from different genes while miRNA only degrade mRNA from their “parent” genes.
piwi-interacting RNAs (piRNAs) combine with piwi proteins to suppression expression and movement of transpons in animal cells via mRNA
CRIPSR-RNAs (ciRNAs) combine with CRISPR to combat foreign DNA.
Chapter 15
Learning Objectives
Draw the general structure of an amino acid (fig 15.5), peptide bonds (fig 15.6), explain the number of common amino acids
Describe primary, secondary, tertiary, and quaternary structure (fig 15.7)
Define codon and know how many possible codons there are
Explain what is meant by the degeneracy of the code
Read the genetic code chart (fig 15.10) – Build a protein from an mRNA strand
Remember that the chart gives the mRNA codon and is written 5’ to 3’
Explain sense codons, synonymous codons, and stop codons
Describe the “wobble” in the code (fig 15.11, fig 15.12); degeneracy of the code
Explain the reading frame of the genetic code; Describe what would happen is there is a shift in reading frame
Describe the steps of translation with key structures/subunits/factors involved (figure 15.21 is a good summary – know the specifics for each step in bacteria, table 15.4 helps)
Amino acid binding tRNAs
Initiation
Elongation
Termination
Explain the differences in translation for prokaryotes and eukaryotes (figure 15.18 and the connecting concepts at the end of this section)
Draw and explain the concept of polyribosomes
Describe and draw simultaneous transcription and translation
Explain post-translational modifications and why they are important
The One Gene, One Enzyme Hypothesis
Discovery
Beetle and Tatum were the first to propose the one gene, one enzyme hypothesis.
This hypothesis states that each gene produces one enzyme or, in some cases, one part of an enzyme.
Research Method:
They used the organism Neurospora to observe the biochemical effects of mutations.
Neurospora is easily cultivated in laboratories and is primarily haploid, allowing direct observation of recessive mutations.
Wild type Neurospora grows on minimal media.
Mutations and Auxotrophs:
Mutations can disrupt fungal growth, leading to auxotrophs, which cannot grow on minimal media but can grow on enriched media containing substances they can no longer synthesize.
Beetle and Tatum irradiated Neurospora spores to induce mutations.
Spores were cultured in various tubes with different media:
One medium contained all necessary biological substances.
These irradiated spores grew into fungi, which reproduced via mitosis.
Identifying Mutations:
Spores were transferred from cultured tubes to minimal medium.
Fungi with auxotrophic mutations did not grow, helping to identify cultures with mutations.
Once a culture was determined to have an auxotrophic mutation, they explored the specific effects:
Mutant strains were transferred to a complete medium, then to tubes containing minimal medium supplemented with various essential biological molecules (e.g., amino acids).
If growth occurred, the specific added substance was identified as the one affected by the mutation.
Multistep Biochemical Pathways:
Adrian Srb and Norman Horowitz applied the outlined procedure to dissect biochemical pathways, specifically focusing on arginine synthesis.
They isolated auxotrophic mutants requiring arginine growth and tested their ability to grow on media supplemented with three compounds: ornithine, citrulline, and arginine.
Results led to mutations categorized in three groups:
Group I: Mutation blocking the synthesis of ornithine, citrulline, and arginine.
Group II: Mutation blocking citrulline and arginine only.
Group II: Mutation preventing arginine synthesis.
Enzymatic Pathway Determination:
This research indicated the sequence precursor → ornithine → citrulline → arginine
Issues with the theory
Although Beetle and Tatum focused on enzymes, it was noted that many genes encode proteins that are not enzymes.
Thus, the broader hypothesis emerged: each gene encodes a protein.
One Gene, One Polypeptide Hypothesis:
Research indicated that many proteins consist of multiple polypeptide chains, each encoded by different genes.
The model was modified further to the one gene, one polypeptide hypothesis.
Composition of Proteins
Amino Acids:
There are 20 common amino acids found in proteins, encoded by three-letter codons.
Each amino acid has a similar structure:
Central carbon atom, amino group, hydrogen atom, carboxyl group, and a distinct R or radical group.
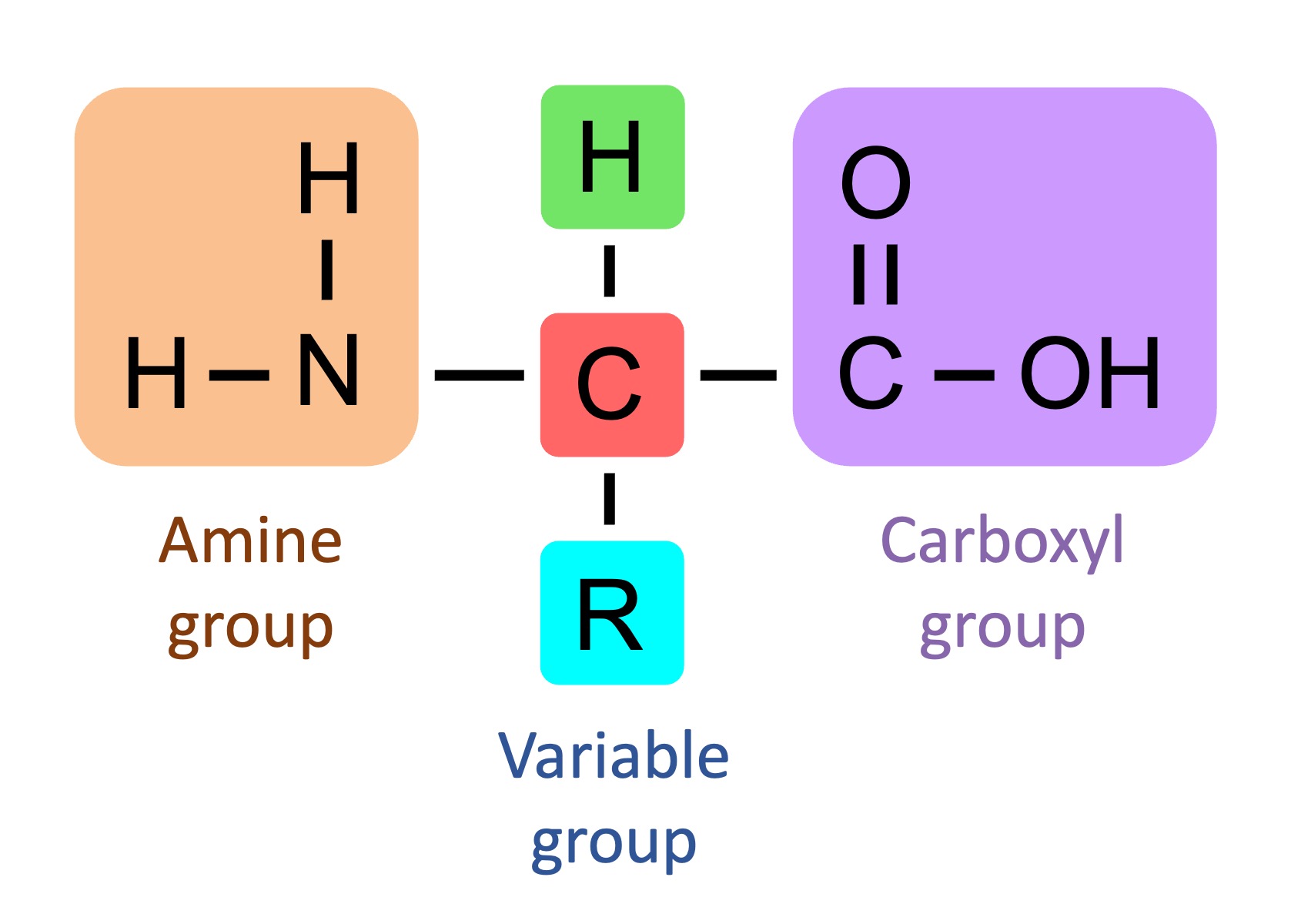
Peptide Bonds
Amino acids link through peptide bonds in polypeptide chains, forming proteins.
Proteins can be composed of one or more polypeptide chains, with a minimum of 50 amino acids and sometimes several thousand.
Polarity of Polypeptides:
Like nucleic acids, polypeptides exhibit polarity:
The amino end has a free amino group (NH_3^+).
The carboxyl end has a free carboxyl group (COO^- ).
Amino acids are added in an N → C or amino → carboxyl.
The carboxyl group is where the next amino acid attaches from
Protein Structure
Levels of Organization:
Proteins have multiple organizational levels:
Primary Structure: Sequence of amino acids.
Secondary Structure: Formed via interactions between neighboring amino acids and hydrogen bonds
Occur at the backbone (carboxyl groups, amine groups, and central carbon)
(ex: alpha helixes and beta-pleated sheets).
Tertiary Structure: Structure formed by R-group interactions and hydrophobic interactions
Quaternary Structure: Association of two or more polypeptide chains.
Domains:
Defined as groups of amino acids forming discrete functional units within a protein, e.g., DNA-binding domains.
The Genetic Code
The question arose regarding how many nucleotides are needed to specify a single amino acid, leading to the conclusion that three nucleotide bases (triplet code) are required to encode for 20 different amino acids. Codons are sets of three nucleotides that code for an amino acid.
Testing the Hypothesis:
Marshall Nirenberg and Johann Matthaei synthesized RNAs via polynucleotide phosphorylase, which does not require a template.
Initial synthetic mRNAs produced were homopolymers (ex: UUU)
Homopolymers - Polymers made of one base
Results of Experiments:
Radioactive labeling in test tubes revealed:
UUU specified phenylalanine.
CCC coded for proline.
AAA encoded lysine.
Additional experiments using random copolymers with varying ratios of nucleotides further defined additional codons.
Nirenberg and colleagues utilized ribosomal bound tRNAs to determine amino acids specified by codons.
Identifying which amino acids were associated with codons through binding and utilizing nitrocellulose filters.
Codons Types
There are 64 total codons
There are 61 sense codons that encode amino acids and three stop codons (UAA, UAG, UGA).
Sense Codons: Codons that code for an amino acid, as opposed to termination
Synonymous Codons: Different codons that code for the same amino acid.
Stop Codons: Codons that do not code for an amino acid and trigger the termination of translation.
UUA
UAG
UGA
The redundancy in the genetic code is termed degeneracy, meaning multiple codons can specify the same amino acid.
Mechanism of tRNA and Codons
tRNA Specificity:
Each tRNA attaches to a single amino acid.
Isoaccepting tRNAs: different tRNAs that accept the same amino acid but have different anticodons.
Wobble Hypothesis:
Discovered by Francis Crick
Wobble Hypothesis: there is flexibility in base-pairing at the third position of the codon.
This flexibility allows tRNA to pair with multiple synonymous codons.
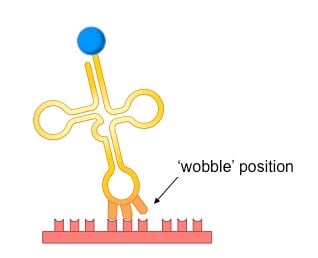
Non-overlapping Code:
The genetic code is non-overlapping, meaning each nucleotide forms part of only one codon.
Initiation codons (mostly AUG) mark the start of translation and specify methionine.
Stop codons do not encode amino acids.
Universal Genetic Code:
Predominantly universal, with exceptions in specific organisms' mitochondrial genes.
Reading Frame
The reading frame defines how an mRNA sequence is grouped into three-nucleotide codons during translation, beginning with the start codon.
A shift in the reading frame, typically caused by insertions or deletions of nucleotides not in multiples of three, alters all downstream codons, leading to an entirely different amino acid sequence and often a non-functional, truncated protein

Polyribosome - When multiple ribosomes work on the same mRNA, found in both Prokaryotes and Eukaryotes
Simultaneous Transcription in Translation - When, in a prokaryote, a ribosome translates mRNA while its still being transcribed by RNA polymerase
Translation Process
Overview:
Translation occurs on ribosomes, beginning at the mRNA's 5' end and moving towards the 3' end.
Protein synthesis proceeds from the amino end to the carboxyl end.
Stages of Translation:
Divided into four major stages:
tRNA Charging: Binding tRNAs to their specific amino acids, facilitated by aminoacyl tRNA synthetases requiring ATP.
Initiation: Assembly of necessary components including mRNA, ribosome subunits, initiation factors, and initiator tRNA.
Elongation: Amino acids are sequentially added to form the growing polypeptide chain.
Termination: The process halts at termination codons, resulting in the release of the newly synthesized polypeptide.
Detailed Stages of Translation
→ Eukaryotic Translation
1. tRNA Charging
Each tRNA is charged with its appropriate amino acid by specific enzymes, aminoacyl tRNA synthetases.
The charging reaction occurs in two steps:
Step 1: Amino acid reacts with ATP, forming aminoacyl-AMP.
Step 2: The amino acid is transferred to the tRNA, releasing AMP.
Errors in tRNA charging are rare, maintaining fidelity through proofreading mechanisms.
2. Initiation Stage
Eukaryotic Initiation
mRNA Transportation: The mRNA needs to travel from the nucleus to the cytoplasm. It does this with the Cap-binding Complex (CBC) which exports mRNA to the cytoplasm and promotes the initial/pioneer round of translation, which is important in checking for mRNA errors.
The small ribosomal subunit, initiation factors, and the initiator tRNA in the P sites form an initiation complex and bind to the 5’ guanine cap
The Kozak sequence containing the AUG start codon is used by the ribosome to identify the start sequence
The initiator tRNA binds to the start codon and te large subunit of the ribosome joins to create a functional ribosome
3. Elongation Stage
Involves three critical steps:
A charged tRNA binds to the A site.
Peptide bonds form between the amino acids in the P and A sites.
Translocation occurs, moving the ribosome to the next codon.
4. Termination Stage
The process terminates when a release factor binds to the stop codon, leading to the release of the polypeptide chain, and all components are dissociated.
→ Prokaryotic Translation
1. tRNA Charging
Each tRNA is charged with its appropriate amino acid by specific enzymes, aminoacyl tRNA synthetases.
The charging reaction occurs in two steps:
Step 1: Amino acid reacts with ATP, forming aminoacyl-AMP.
Step 2: The amino acid is transferred to the tRNA, releasing AMP.
Errors in tRNA charging are rare, maintaining fidelity through proofreading mechanisms.
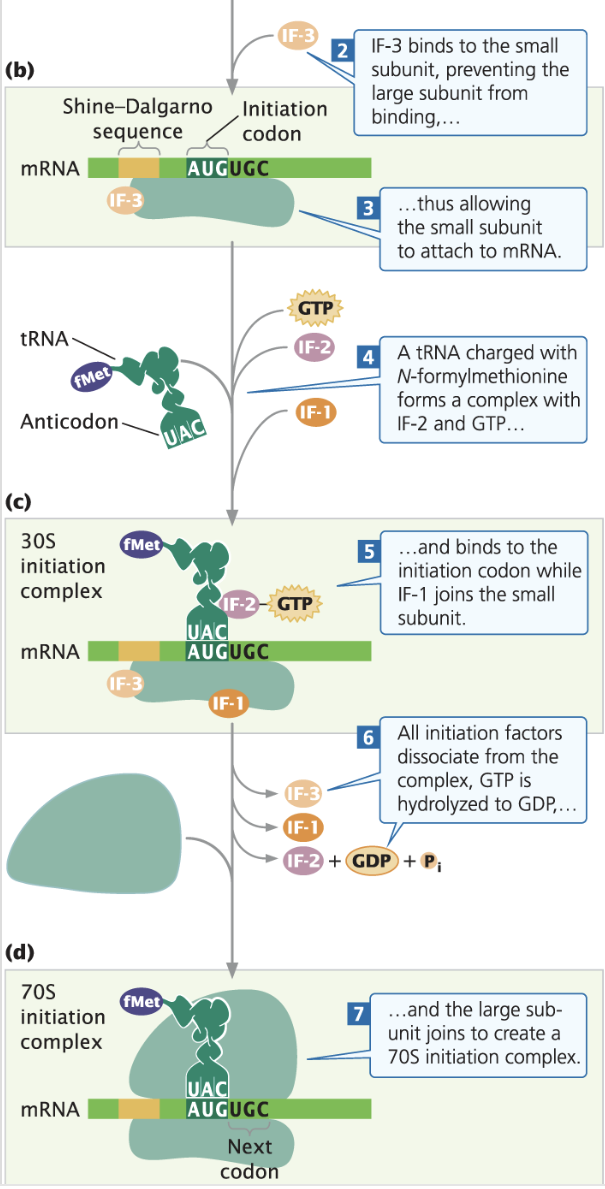
Bacterial Initiation
IF-3 binds to the small subunit (30s) to prevent the large subunit (50s) from attaching to the small unit early
Then mRNA’s Shine-Dalgorno sequence binds to the small subunit of the ribosome
Then initiation tRNA, charged with N-formylmethionine and combined with IF-2 and GTP base pairs with the start codon in the p site.
IF-3, IF-2, and IF-1 are unbound using the energy from GTP, turning it into GDP
The large subunit attaches, forming the complete ribosome (70S)
Prokaryotic elongation
An Elongation Factor Tu (EF-TU) and GTP help a charged tRNA bind into the A site of the ribosome, and the anticodon binds to the codons
Elongation Factor Ts (EF-Ts) help regenerate the GTP and EF-tu
The ribosome translocates down the mRNA, using Elongation Factor G (EF-G)
Prokaryotic termination
Once the ribosome reaches the stop codon, instead of tRNA Release Factor 1 (RF-1) or 2 (RF-2) bind to the codon, causing the polypeptide chain to be cleaved and released
Release Factor 3 (RF-3) and GTP bind with the ribosome, releasing RF-1 & RF-2
Quality Control Mechanisms
Quality control in translation prevents errors during synthesis and involves several mechanisms such as:
Nonsense-mediated mRNA decay (NMD) to eliminate mRNAs with unexpected termination codons.
Transfer messenger RNAs (tmRNAs) in bacteria can recycle stalled ribosomes.
Post-Translational Modifications
Newly synthesized proteins often undergo various modifications:
Cleavage of precursor proteins, removal of amino acid sequences, signal sequences for localization, or addition of functional groups (e.g., acetylation).
These modifications let mRNA create more than one protein
Antibiotics and Translation
Antibiotics target translation selectively, affecting bacterial ribosomes while sparing eukaryotic ones.
Classes of antibiotics
Tetracyclines: Block tRNA entry to the ribosome A site.
Streptomycin: Prevents initiation of translation.
Erythromycin: Blocks translocation of ribosomes.