DNA replication
DNA replication: a semi conservative process in which DNA makes a copy of itself.
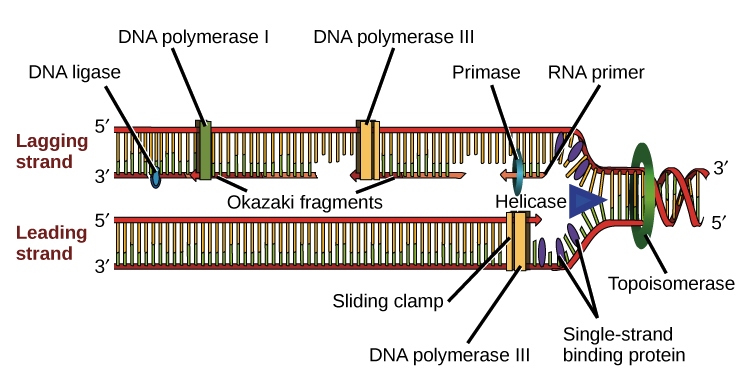 1. Replication Fork Formation
1. Replication Fork Formation
Unwind DNA by using the protein helicase
Helicase binds to a location known as the origin of replication to unwind the double stranded DNA
DNA helicase disrupts the hydrogen bonds between base pairs to separate the strands into Y shape known as the replication fork.
The single stranding Binds proteins attaches to the single strands to stabilize the bases
Replication Fork: bidirectional (the leading strand goes 3’ to 5’ while the lagging strand is oriented with vice versa).
Topoisomerase works at the region ahead of the replication fork to prevent supercoiling.
Fork of replication: location of DNA helicase
*The unwinding process introduces positive supercoils where an enzyme called DNA gyrase/topoisomerase has to introduce negative supercoils decreasing the number of positive supercoils.
2. Primer Binding
These single stranded DNA complementary molecules are exposed to their own processes cataylzed by the enzyme primase/RNA polymerase.
RNA polymerase binds to the single stranded molecules at 3’ and add sets of new nucleotides known as RNA primer.
Acts as a signal to DNA polymerase
RNA primer: a sequence of nucleotides to activate the synthesis of daughter strands
DNA replication: Elongation
DNA polymerase bind to the primer region and add indiviudal nucleotides to leading and lagging strands.
DNA polymerase catalyzes the formation of the phosphodiester bond by adding deoxynucleoside triphosphate into the growing polynucleotide chain
It takes the free nucleotides and attaches the nucleotides to form a phosphodiester linkage releasing pyrophosphoric acid.
DNA polymerase can only read the parent strand and can only synthesize the daughter strand in the 5-3 direction, creating nucleotides beginning with 5 prime and ending with 3’ (leading strand)
leading strand forms continuosly in the same motion as the fork
In eukaryotic cells, polymerases alpha, delta, and epsilon are the primary polymerases involved in DNA replication. Because replication proceeds in the 5' to 3' direction on the leading strand, the newly formed strand is continuous.
Many RNA primers are formed to synthesize the lagging strand
nucleotides are formed in the backward direction
moving backwards from the helicase, reversed
ensures that the DNA polymerase forms in the 5’ to 3’ direction.
Okazaki fragments: name given to the pieces in the piece wise fashion, the gaps formed between the lagging strands.
Removal of the gaps:
Remove the primers and replace them with proper nucleotides with the enzyme DNA ligasae
Connects the okazaki fragements by creating phosphodiester linkage.
4. Termination:
The terminus ultilization substance binds to the terminator sequence to stop DNA polymerase.