Cell Structure Final Exam
1/129
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
130 Terms
primary transcript
a newly produced RNA molecule
RNA processing
necessary chemical modification for newly produced RNA before it can function in the cell, generates a “mature” RNA product
mRNA processing in eukaryotes
involves capping, addition of Poly (A), and removal of introns
why bacteria are translated as they are transcribed
because there is no nuclear membrane
heterogeneous nuclear RNA (hnRNA)
very long primary transcripts that are usually 2000- 20,000 nucleotides, a mixture of mRNA and pre-mRNA
pre-mRNA
precursors to mRNA, processed by removal of sequences and addition of 5’ caps and 3’ tails
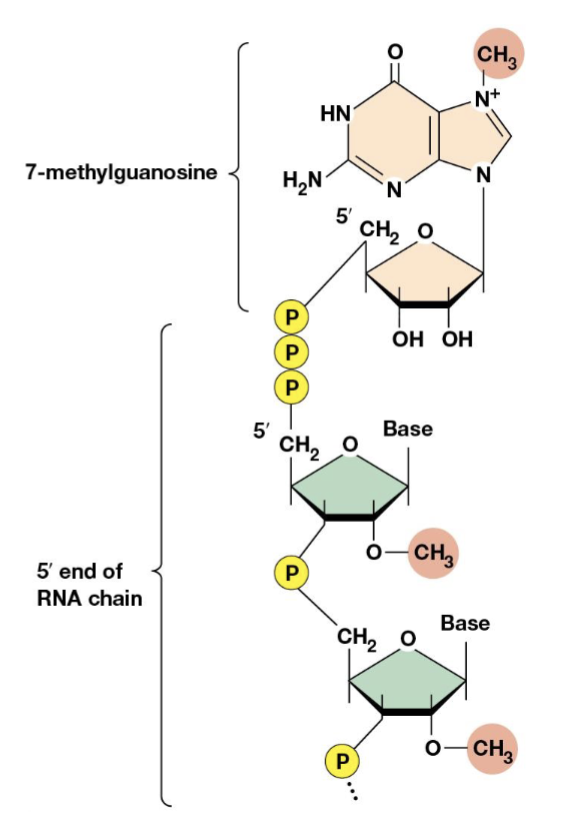
5’ cap
a guanosine that is methlated at position 7 of the purine ring, bound to the RNA by 5’ to 5’ linkage
contributes to mRNA stability by protecting the RNA from nucleases, plays a role in positioning the RNA on the ribosome for initiation of translation
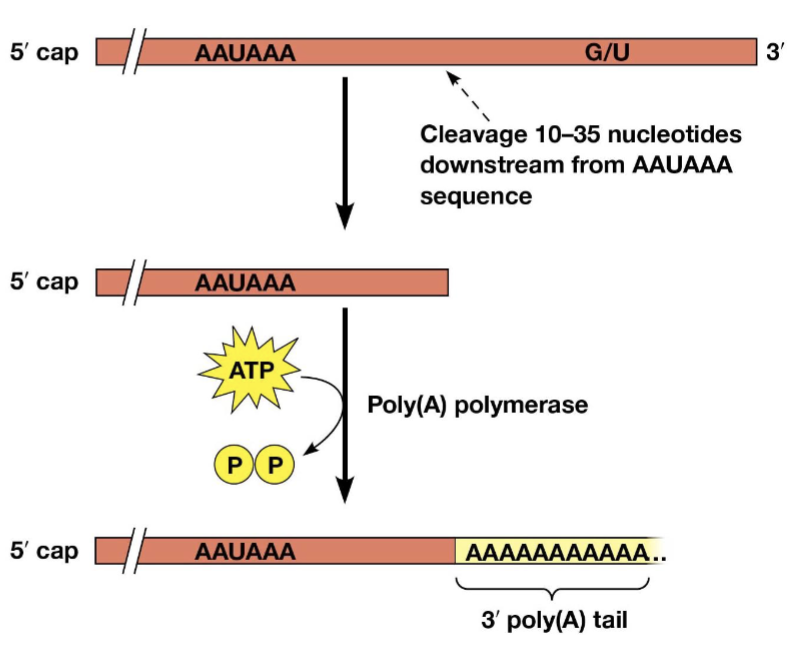
poly(A) tail
ranges from 50 to 250 nucleotides long
protects the mRNA from nuclease attack, required for export of the transcript to the cytoplasm, may also help ribosomes recognize and bind to mRNAs
poly(A) polymerase
adds the poly(A) tail
AAUAAA
a signal for addition of the poly(A) tail, located just upstream of the polyadenylation site
exons
sequences that appear in the final mRNA
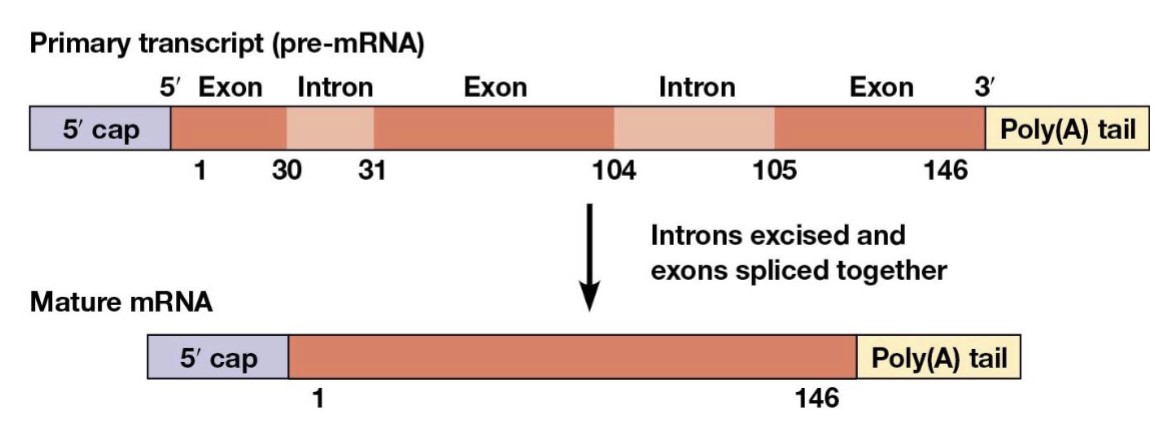
introns
present in most protein-coding genes of multicellular eukaryotes
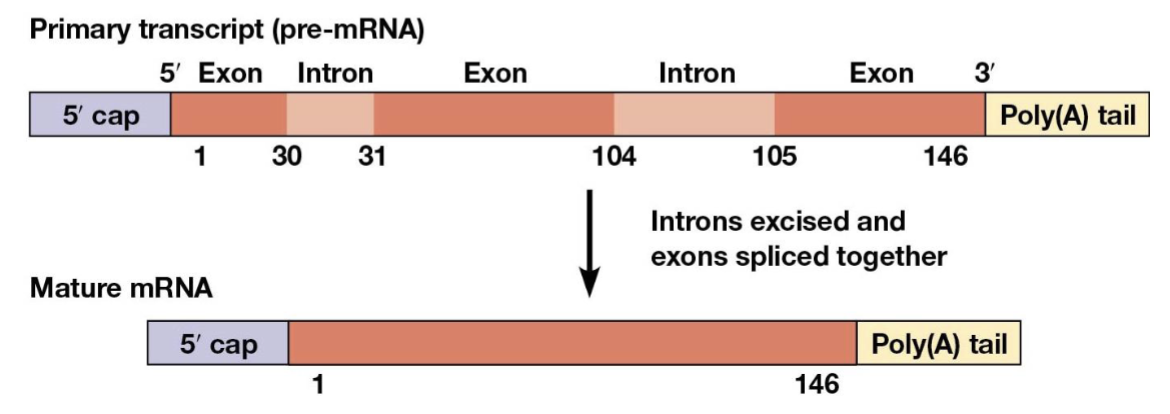
RNA splicing
the process of removing introns and joining the exons
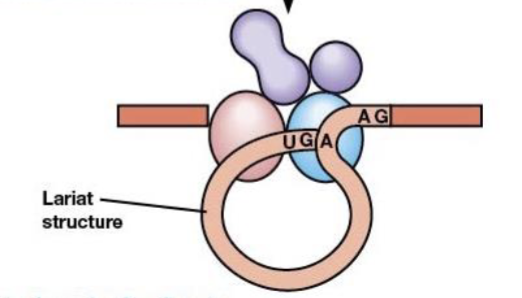
branch point
one additional sequence near the 3’ end of the intron, contains an A residue
splice sites
specific nucleotide sequences at the boundaries of introns and exons in a pre-mRNA that are recognizd by the splicing machinery to remove introns and join exons
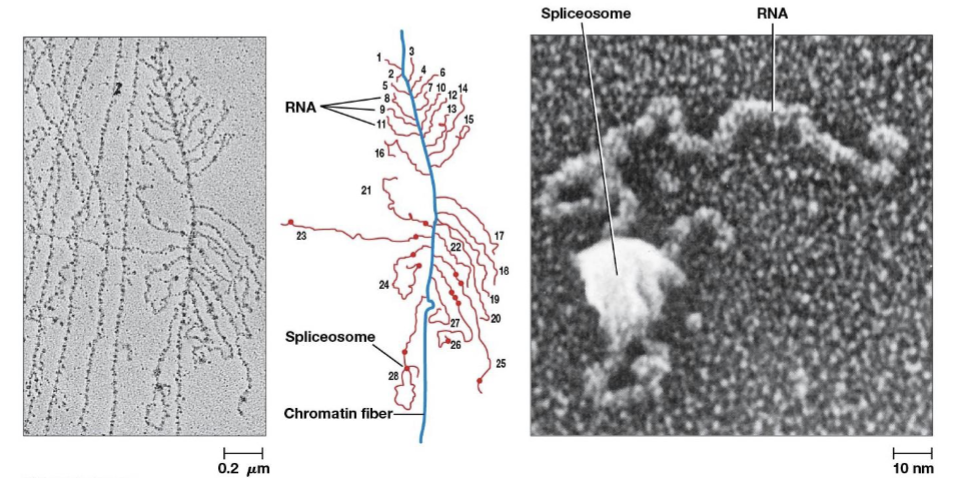
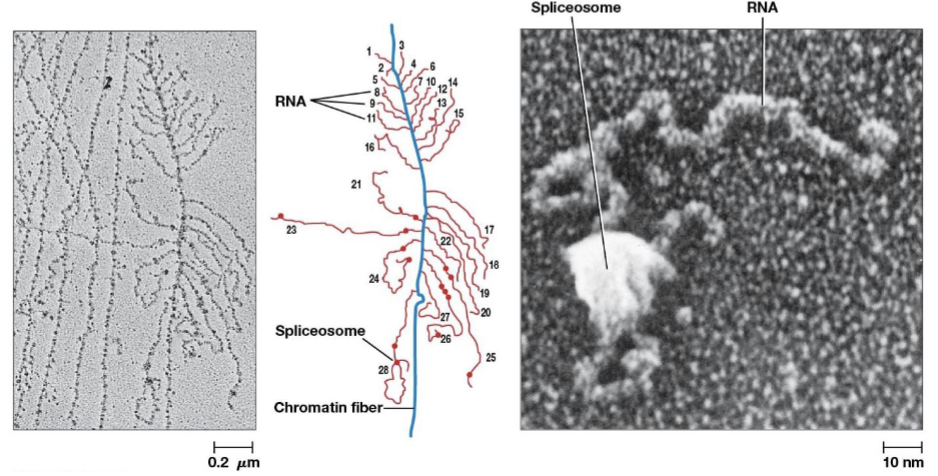
spliceosomes
catalyze intron removal, consists of 5 types of RNA and more than 200 proteins
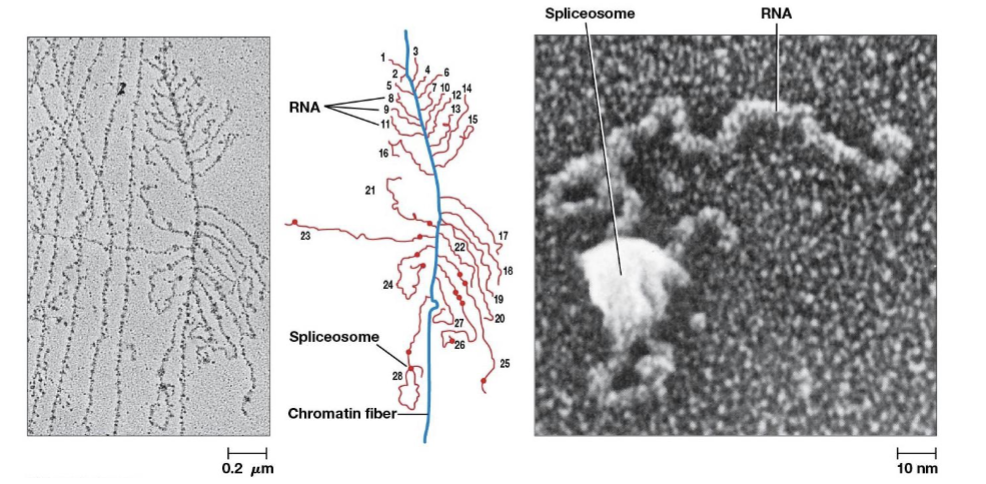
small nuclear ribonuclear protein complexes (snRNPs)
RNA-protein complexes in the cell nucleus that are the building blocks of the spliceosome
snRNA
non-coding RNA molecules in the nucleus of eukaryotic cells that primarily function in pre-mRNA splicing
spliceosome assembly
binding of U1 to pre-mRNA
U2 snRNP binds to pre-mRNA
U4/U6 and U5 snRNPs bind
RNA is cleaved at 5’ splice site and lariat structure is formed
RNA is cleaved at 3’ splice site, exons are joined, and an EFC is added
excised intron is degraded
they are assembled by sequential binding of snRNPs to pre-mRNA
lariat
formed from excised introns during RNA splicing
exon junction complex (EJC)
required for efficient export of the mRNA, influence regulatory events like degradation of mRNA
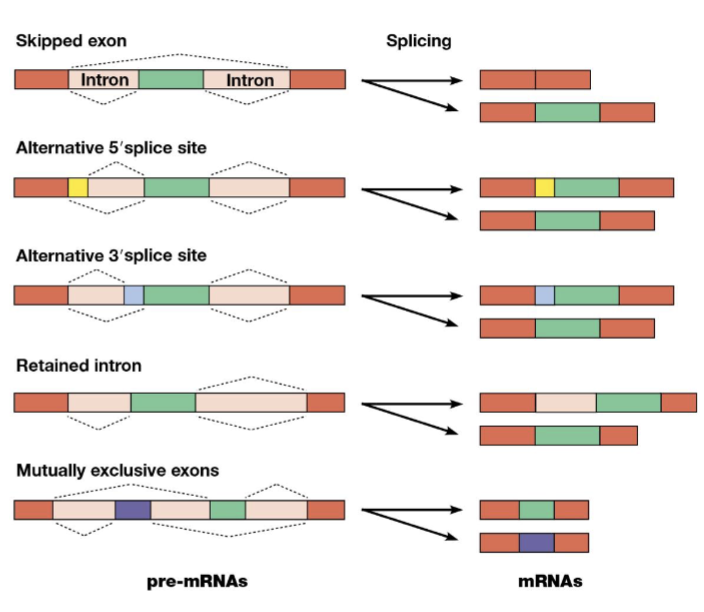
alternative splicing
a process where a single gene can produce multiple different proteins by joining different combination of exons from the initial pre-mRNA transcript
nuclear RNA export factor (NXF 1)
interacts with cap-binding complex and exon junction complex so the mRNA cargo can exit the nucleus and enter the cytosol
cap-binding complex (CBC)
proteins that bind to the mRNA 5’ cap
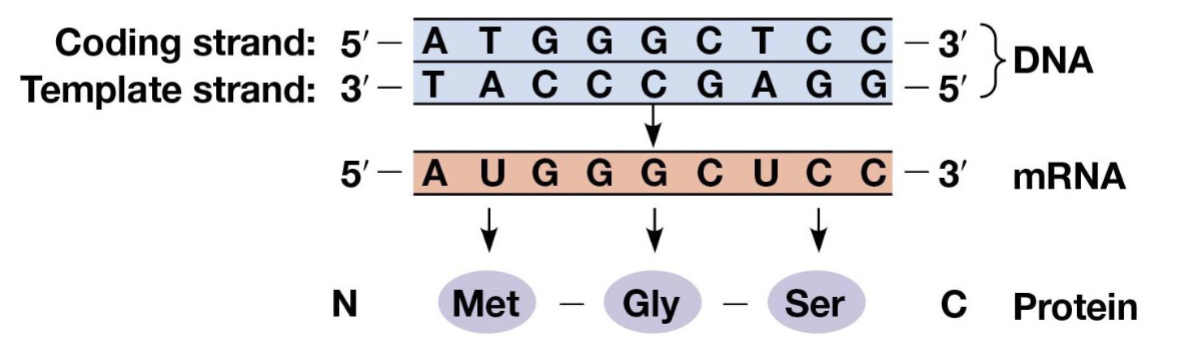
triplet code
combinations of three bases specify amino acids, the genetic code
degenerate code
particular amino acids can be specified by more than one triplet
nonoverlapping
the reading frame advances three nucleotides at a time
codons
RNA triplets, read and written in a 5’ to 3’ manner
start codon
AUG
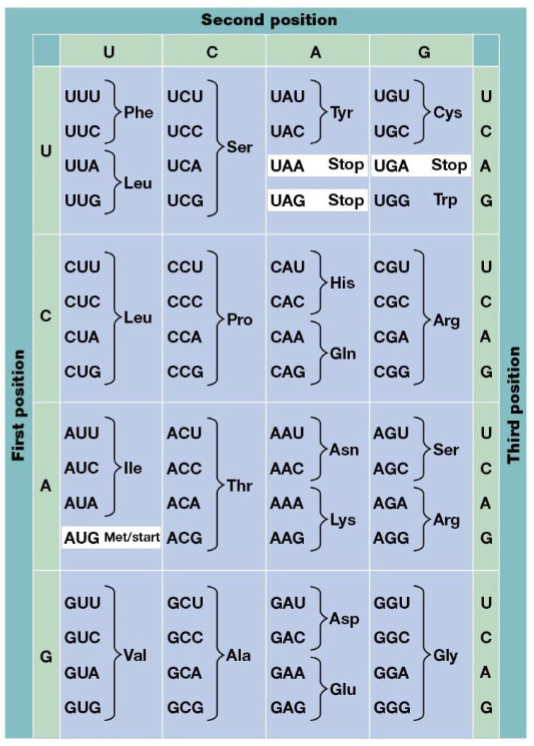
stop codons
UAA, UAG, UGA
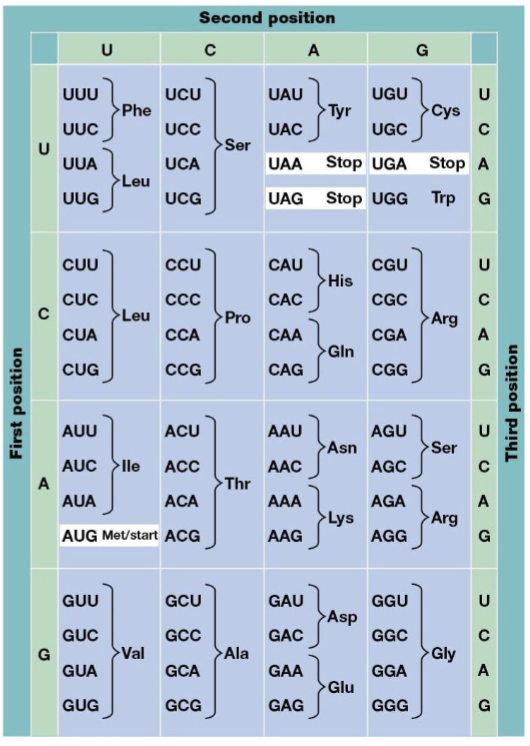
unambiguous
every codon has only one meaning
degenerate
multiple different three-nucleotide codons can specify the same single amino acid
ribosomes
carry out the process of polypeptide synthesis
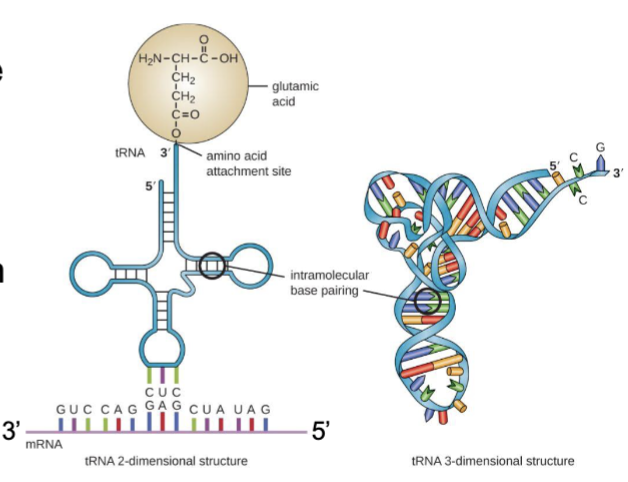
tRNA molecules
align the amino acids in the correct order, an adaptor that binds both a specific amino acid and the mRNA sequences that specify the amino acid
each one is linked to its amino acid by an ester bond
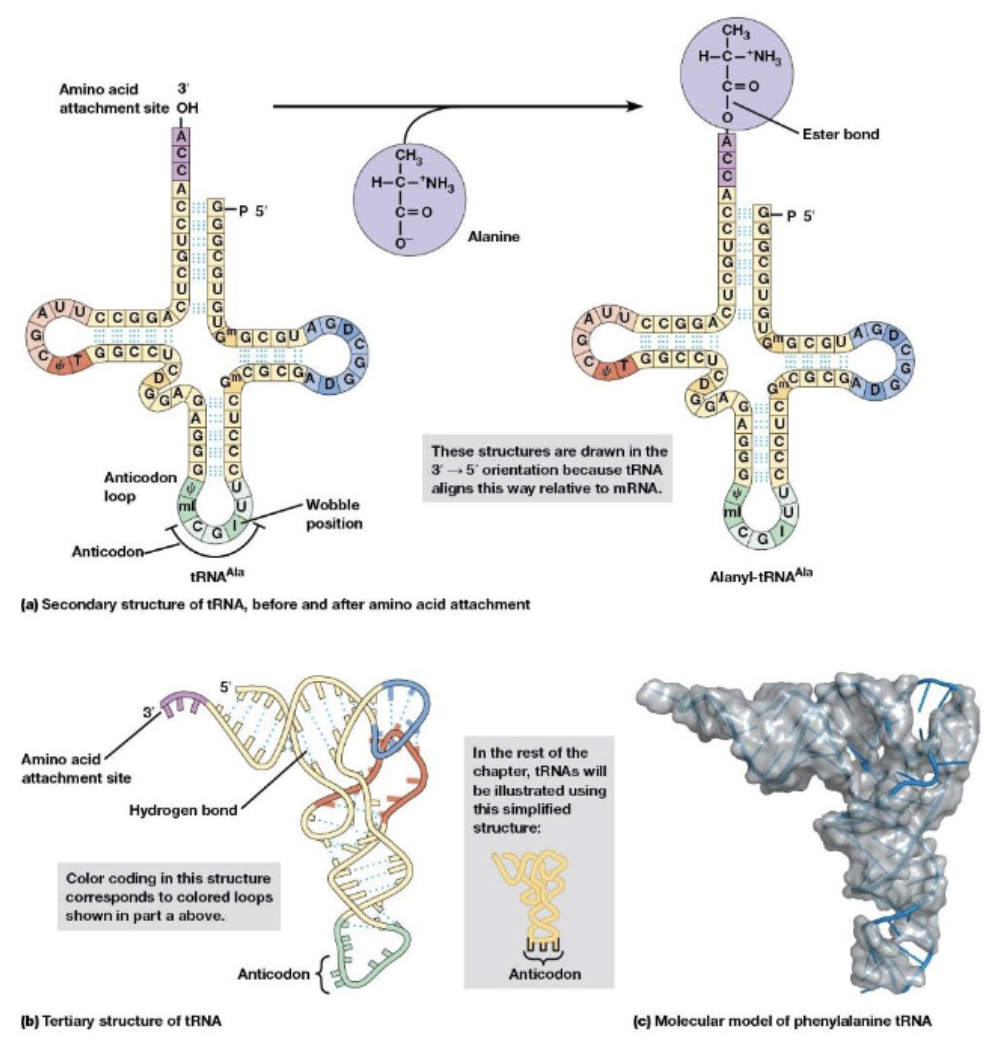
aminoacyl-tRNA synthetases
attach amino acids to their appropriate tRNA molecules, the tRNA is called charged and the amino acid is called activated, there are 20 different ones in cells
mRNA
encode the amino acid sequence information
protein factors
facilitate some of the steps of translation
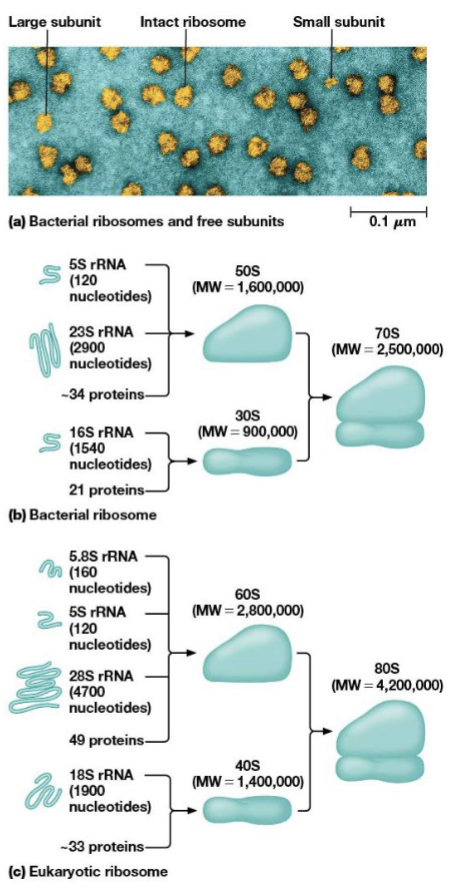
ribsosome structure
built from dissociable subunits, the large and small subunits
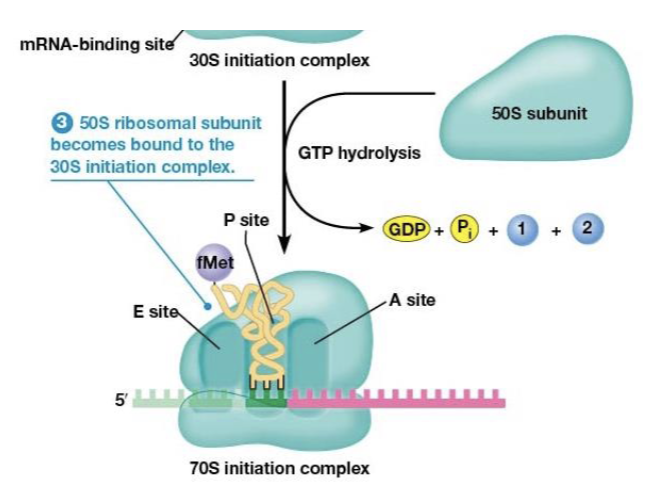
ribosome site for protein synthesis
mRNA-binding site, A, P, and E sites
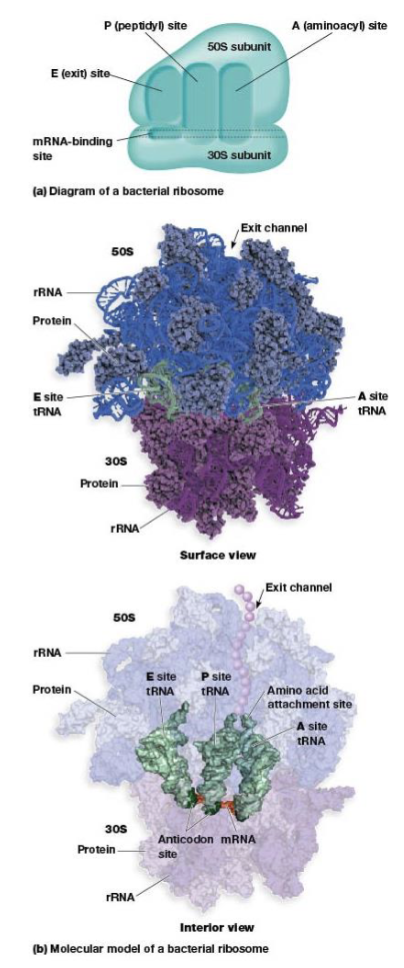
A (aminoacyl) site
binds tRNA with attached amino acid
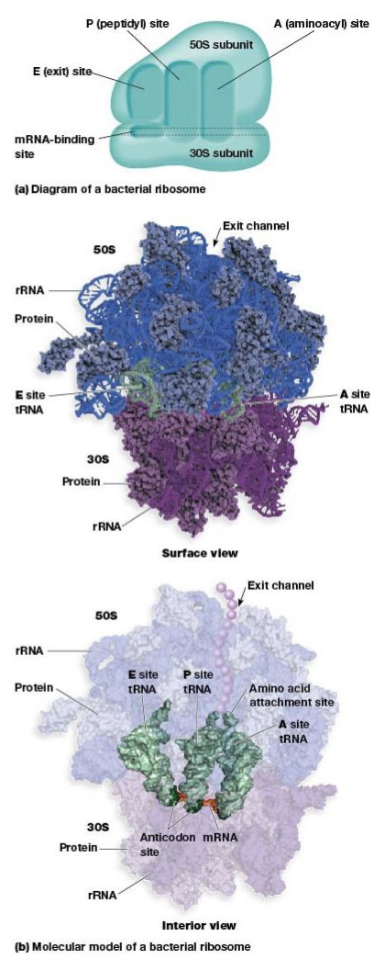
P (peptidyl) site
where the tRNA carrying the growing peptide resides
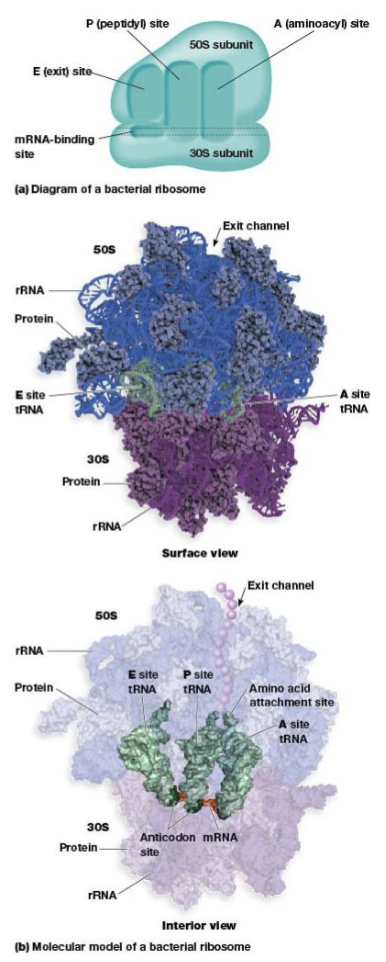
E (exit) site
from which tRNAs leave the ribosome after discharging their amino acid
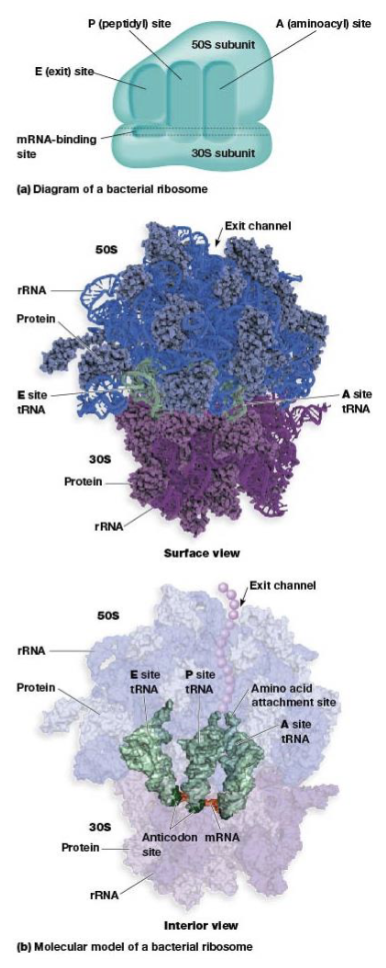
anticodons
permit tRNA molecules to recognize codons in mRNA by complementary base pairing
where translation of the polypeptide begins
N-terminus
initiation factors (IF1, IF2, and IF3)
bind to the small (30S) ribosomal subunit
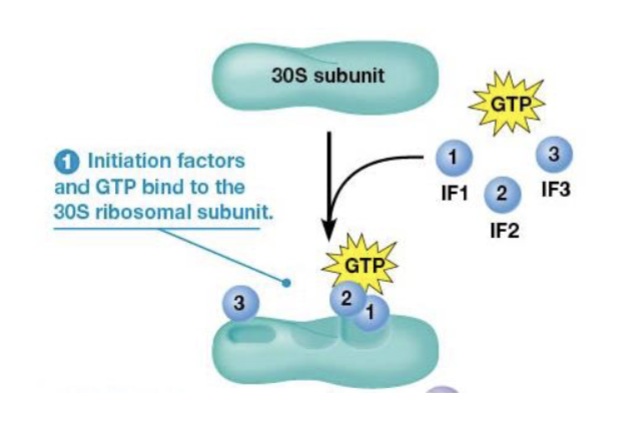
Shine-Dalgarno sequence
consists of 3-9 purines located upstream of the start, the mRNA binds in the proper orientation due to this sequence
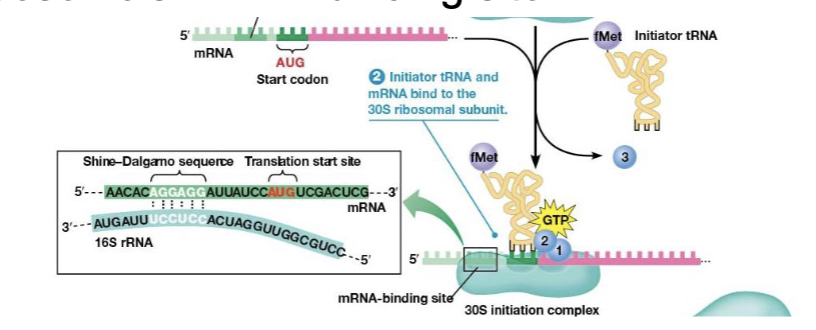
N-formylmethionine (fMet)
used to initiate translation; only its carboxyl group can bind to another amino acid
initiator tRNA
binds the P site of the small subunit by action of IF2 plus GTP
70S initiation complex
formed when IF3 is released and the 30S initiation complex binds to the large (50S) subunit
internal ribosome entry sequence (IRES)
a sequence of mRNA that allows ribosomes to initiate protein synthesis internally, independent of the 5’ cap
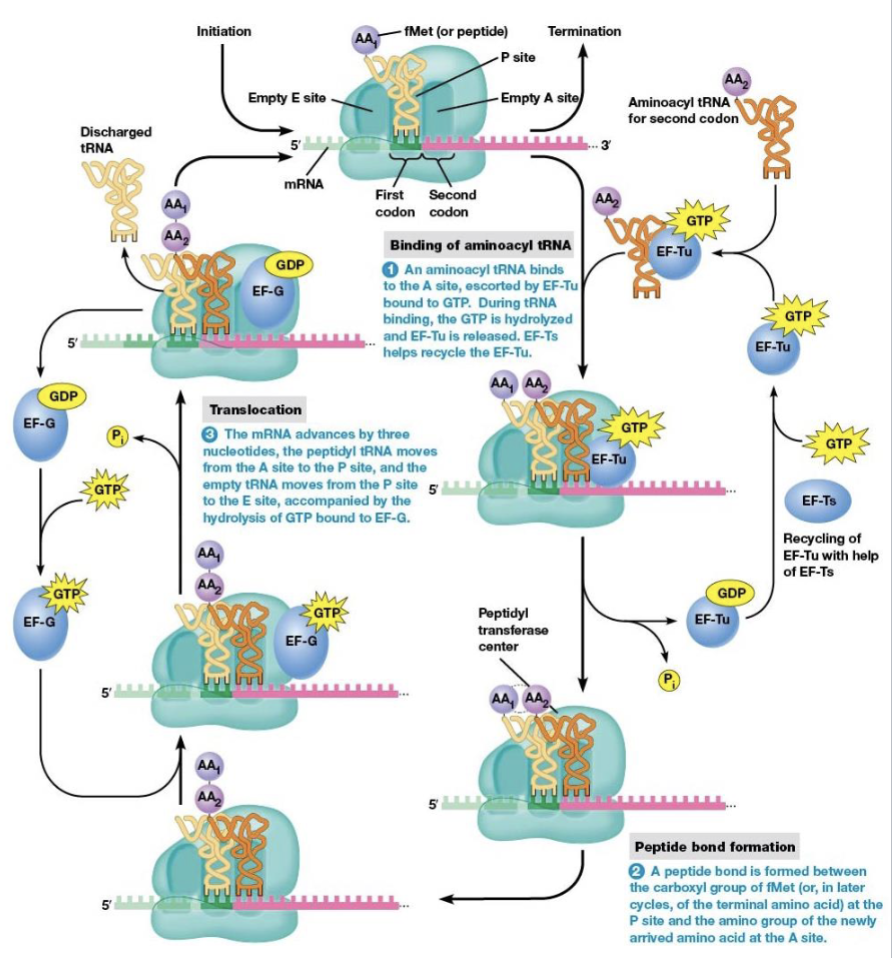
steps of elongation
binding of aminoacyl tRNA to the ribosome
peptide bond formation
translocation
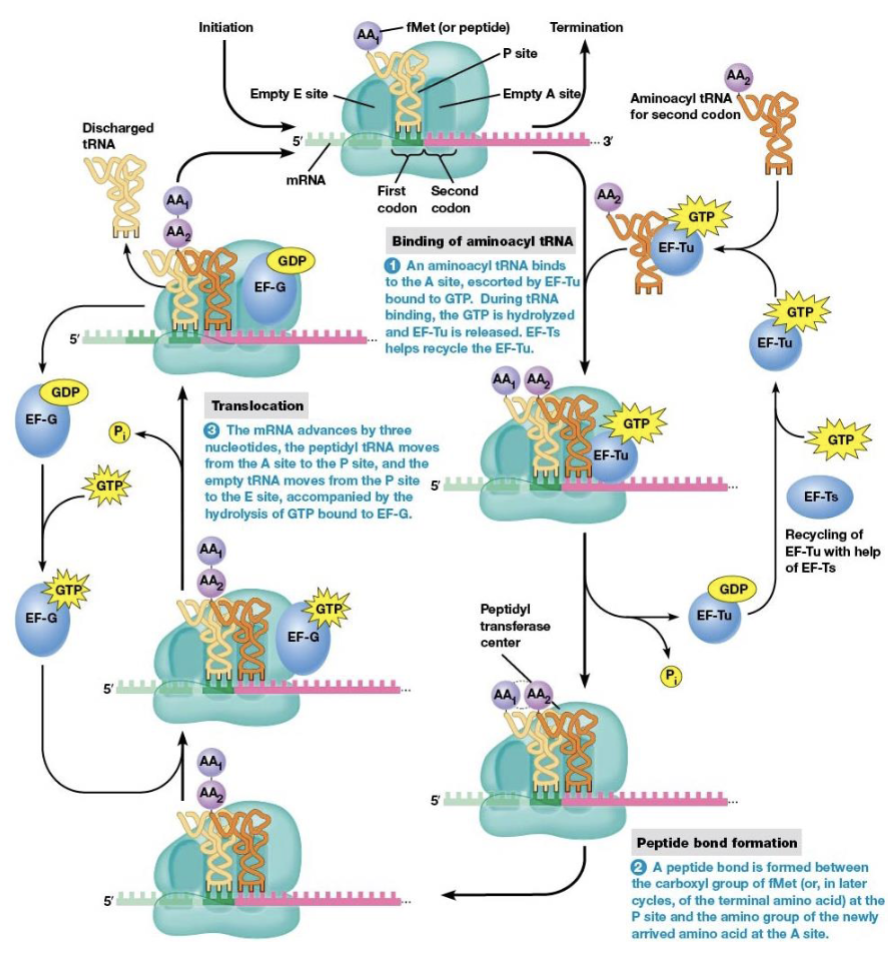
binding of aminoacyl tRNA to the ribosome
brings a new amino acid into position to be joined to the peptide chain
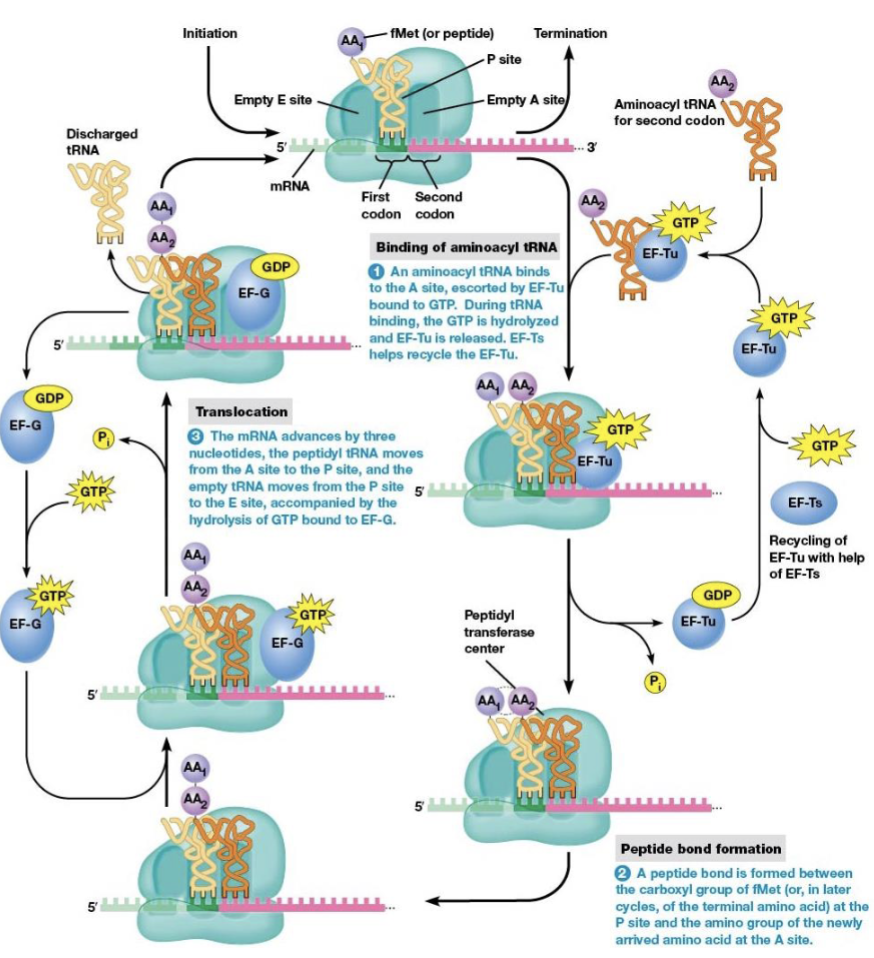
peptide bond formation
links the amino acid to the growing polypeptide
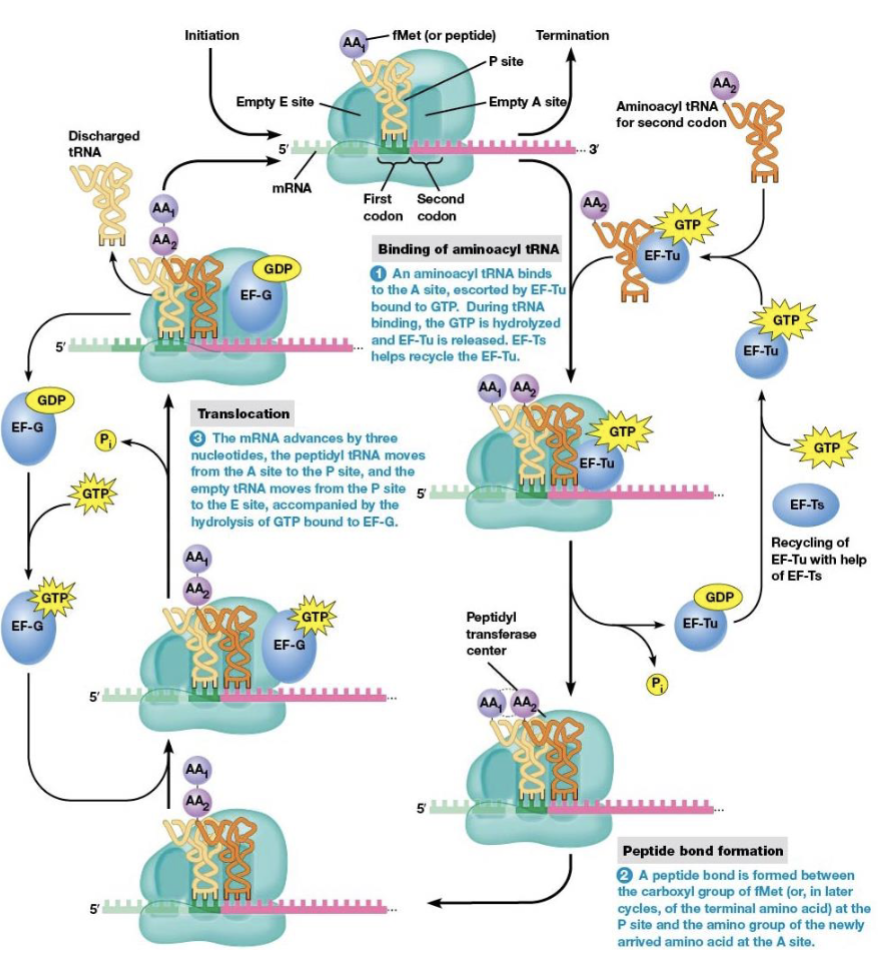
elongation factors
EF-Tu and EF-Ts are necessary for elongation to begin
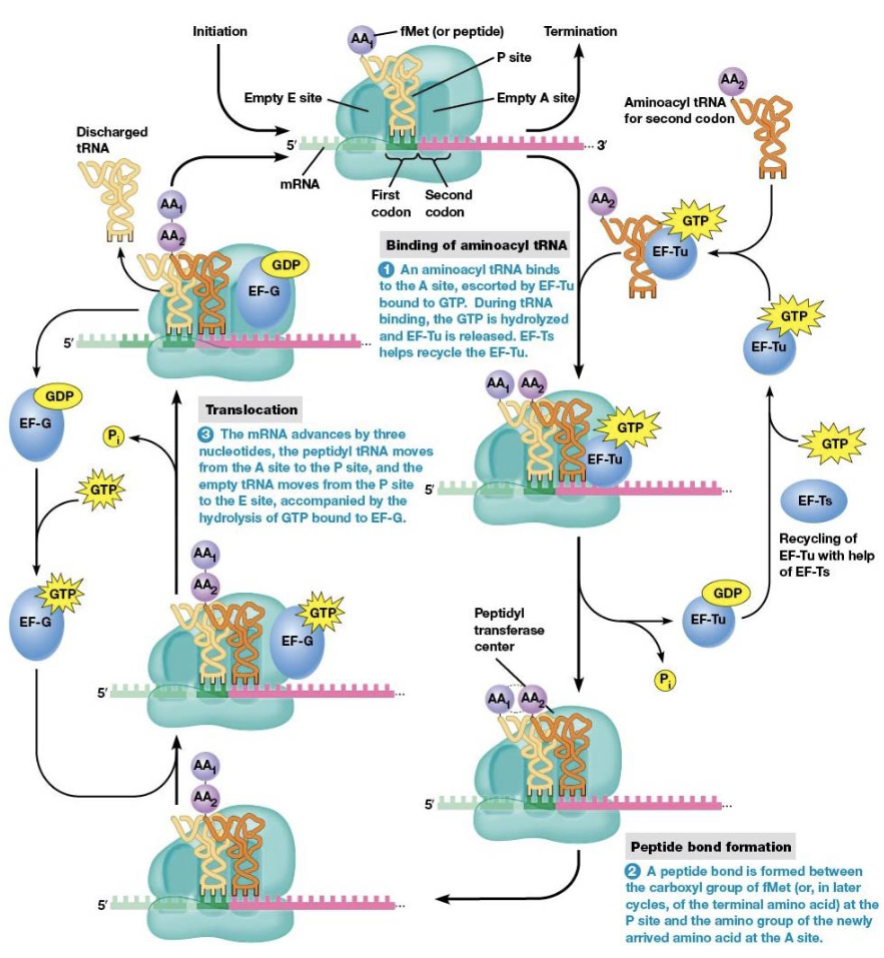
location of the start codon as elongation begins
P site (the next codon is at the A site)
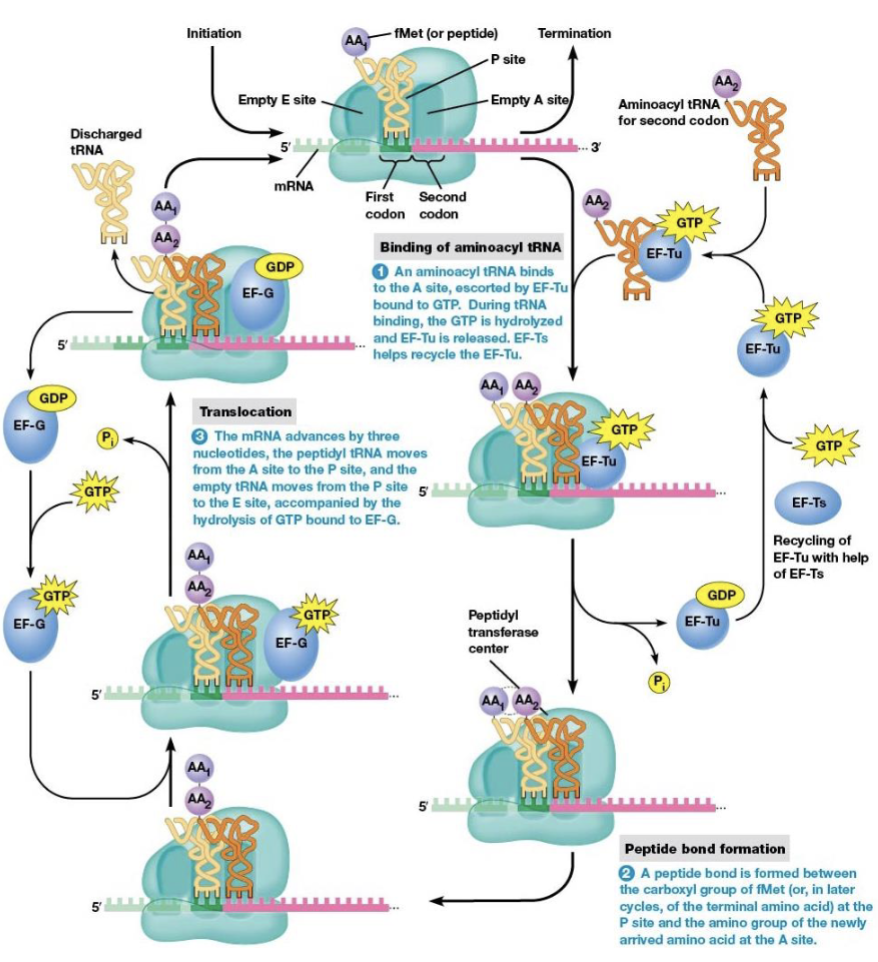
what begins elongation
tRNA with an anticodon complementary to the second codon binds the A site
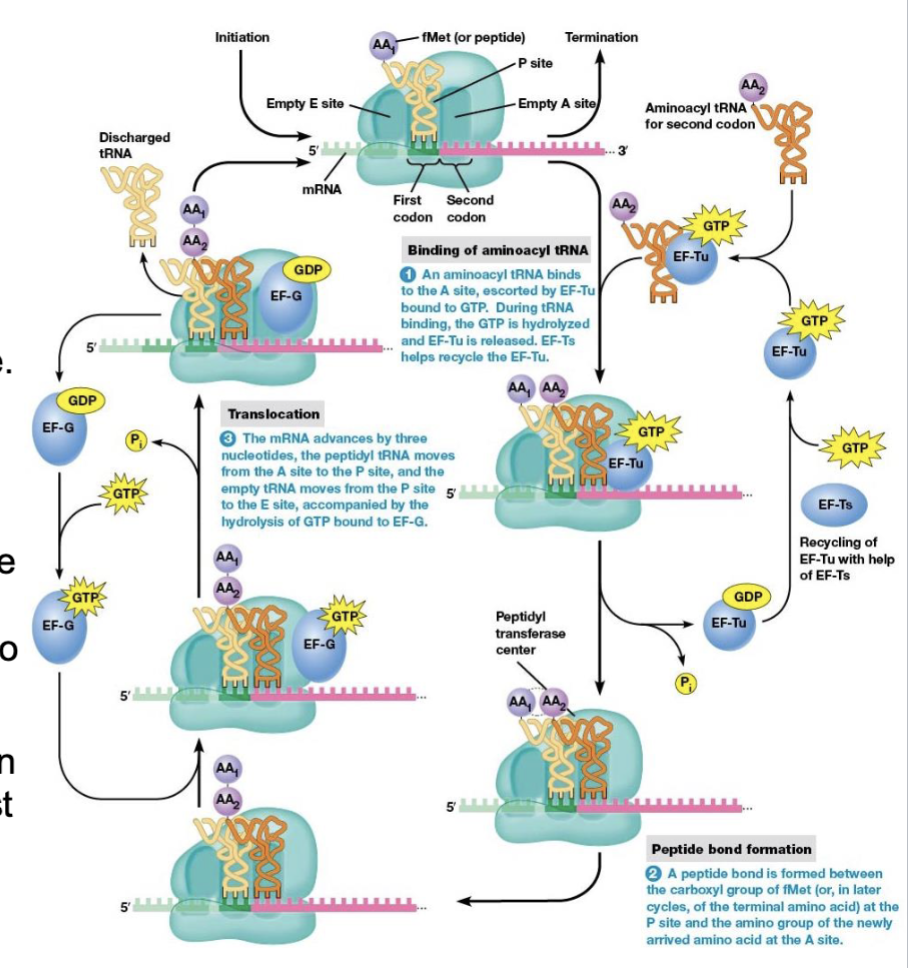
translocation
the process in amino acid synthesis where the ribosome moves along the mRNA strand by one codon after a peptide bond is formed
protein release factors
recognize stop codons and stop translation, bind mRNA stop codons at the A site
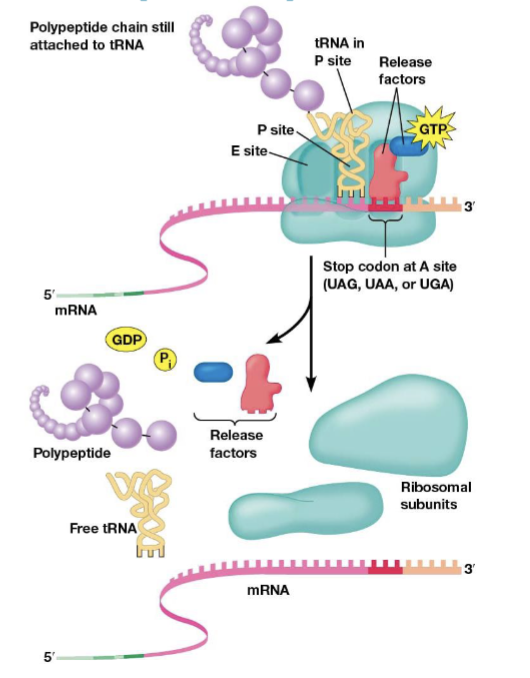
molecular mimicry
the phenomenon where the shape of protein release factors is similar to tRNAs
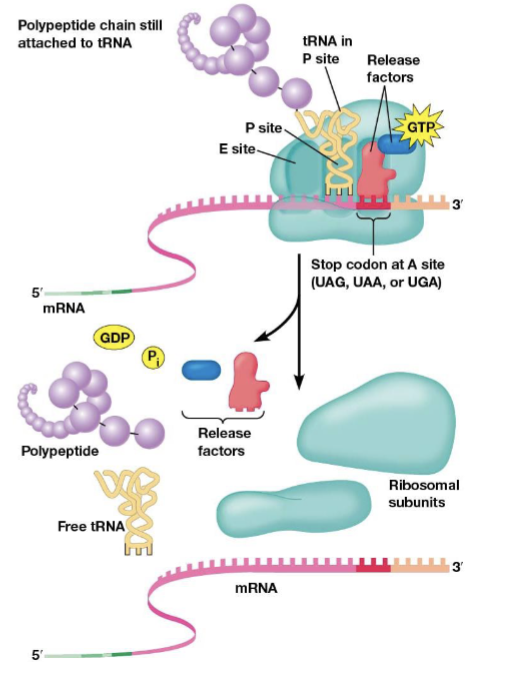
mutation
any change in nucleotide sequence of a genome
base-pair substitution
can alter a codon (this is the case in sickle-cell anemia)
missense or nonsynonymous mutation
encodes for the “wrong” amino acid (changes an amino acid)
nonstop mutation
when a base-pair substitution alters a stop codon to an amino acid codon
nonsense mutation
the mutation changes an amino acid codon to a stop codon
indels
genetic mutations that result from the insertion or deletion of one or more DNA base pairs in a DNA sequence
frameshift mutation
caused by the insertion or deletion of nucleotides in a DNA sequence, but not a multiple of three
silent or synonymous mutations
affect the third base of the codon and results in no change of amino acid
nonsense-mediated decay
used in eukaryotic cells to destroy mRNAs containing premature stop codons, mammals use the EJC to do this
nonstop decay
an RNA-degrading enzyme binds the empty A site of the ribosome and degrades the defective mRNA
result of an mRNA lacking a stop codon
translation becomes stalled
tmRNA
an RNA that has a tRNA domain that gets charged, and an mRNA domain that has a stop codon
posttranslational modification
in bacteria, N-formyl group is removed and the methionine at the N-terminus is often removed
in eukaryotes, the methionine at the N-terminus is often released
central nervous system (CNS)
consists of the brain and spinal cord
peripheral nervous system (PNS)
comprises other sensory or motor components
main types of cells in the nervous system
neurons
glilal cells
neruons
send and receive electrical impulses (nerve impulses)
sensory neurons
responsible for the detection of stimuli
motor neurons
transmit signals from the CNS to the muscles and glands, have multiple branched dendrites and a single long axon
interneurons
signals and transmits information between parts of the nervous system
glilal cells
encompass a variety of cell types, are the most abundant type of cell in the CNS
microglia
fight infections and remove debris
oligodendrites and schwann cells
form the insulating myelin sheath around neurons of the CNS and peripheral nerves
astrocytes
control access of blood-borne components into the extracellular fluid around the nerve cells, forming the blood-brain barrier
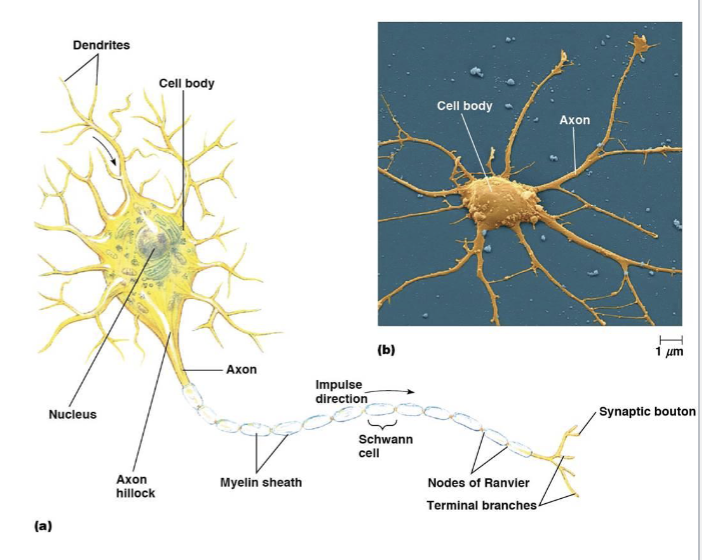
neuron cell body
includes the nucleus and other endomembrane components
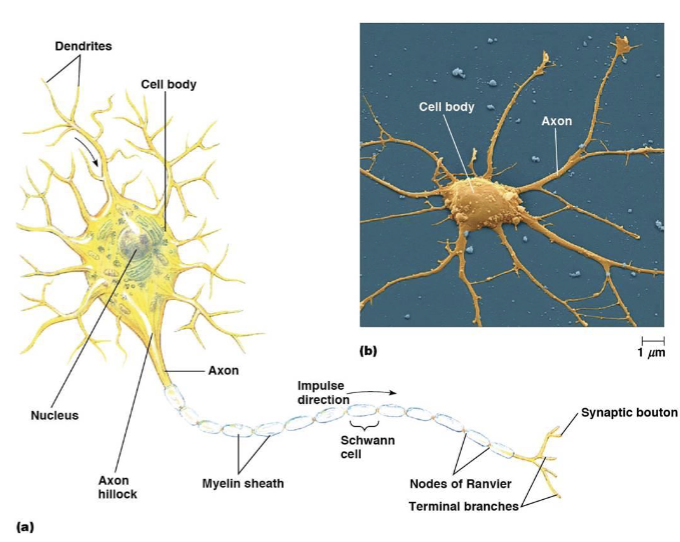
dendrites
receive signals
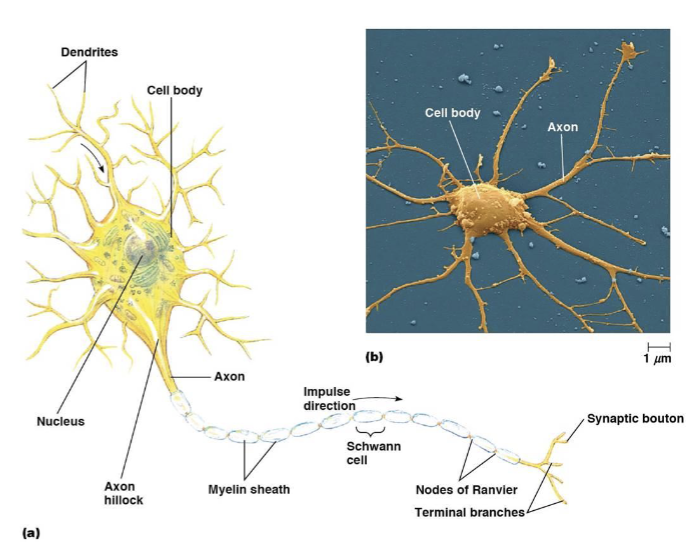
axons
conduct signals
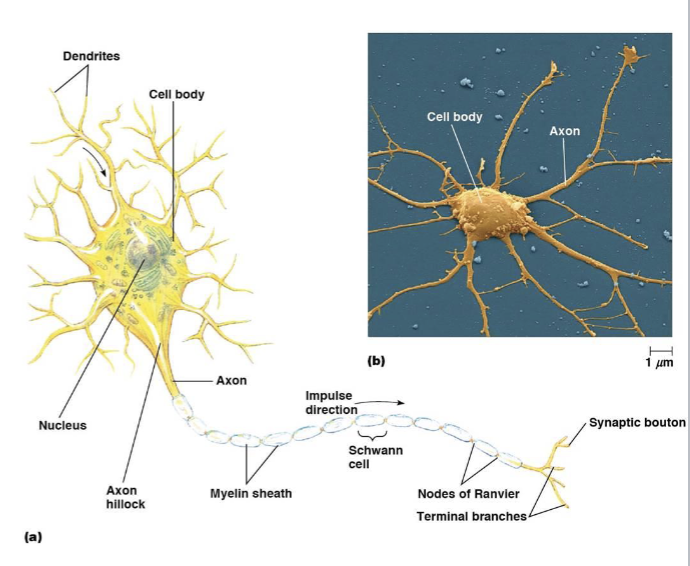
myelin sheath
a fatty, insulating layer that surrounds nerve axons, allowing electrical impulses to be transmitted quickly and efficiently down the nerve
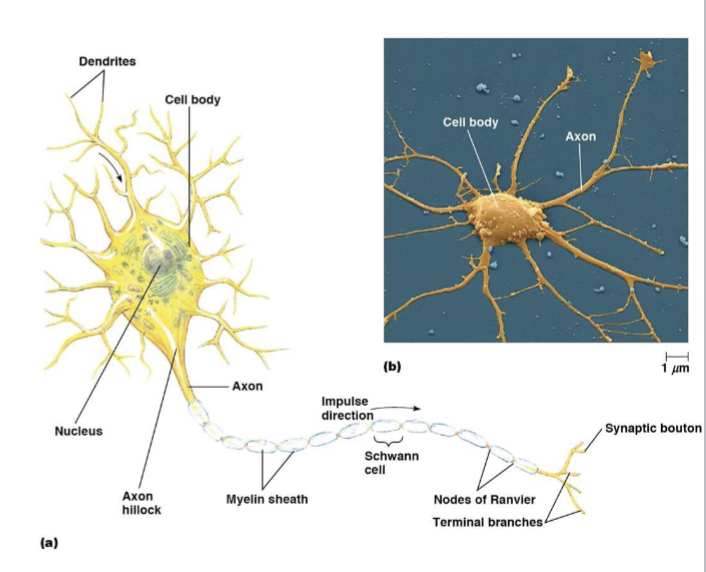
nodes of Ranvier
a gap in the myelin sheath of a nerve
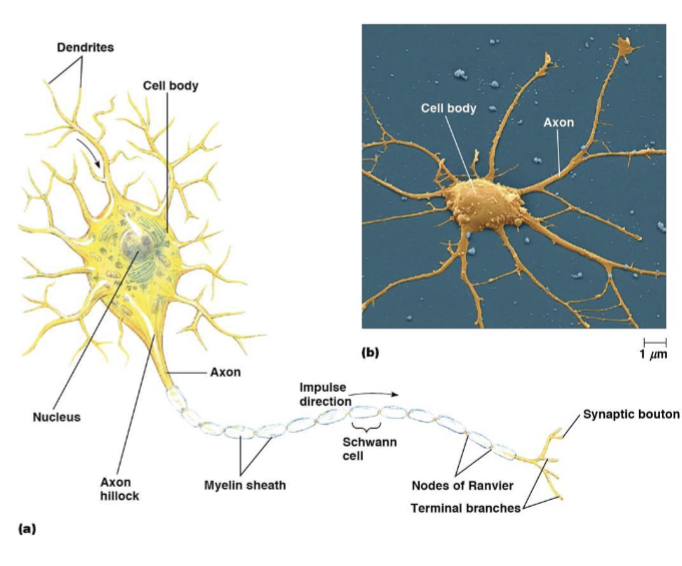
synaptic boutons
transmit the signal to the next cell, a neuron, muscle, or gland
purpose of myelination
it decreases the ability of the neuronal membrane to retail electrical charge (decreases capacitance), allows nerve impulses to spread farther and faster
synapse
the junction between a nerve cell, gland, or muscle cell
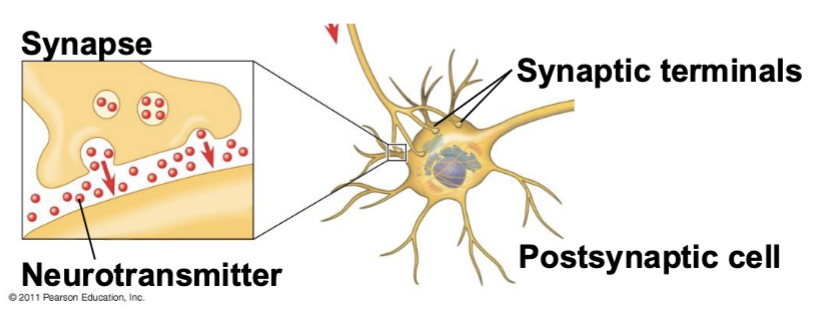
neurotransmitters
chemical messengers that carry information across the synapse
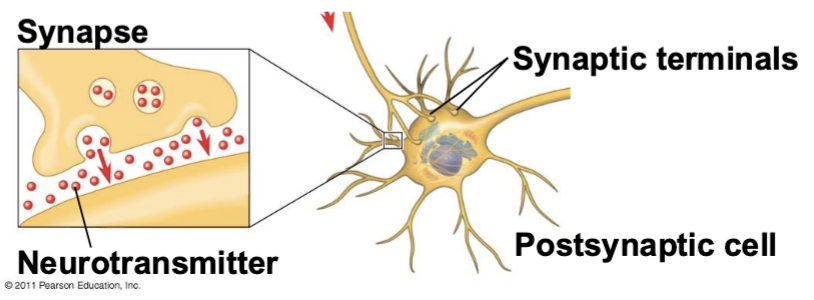
presynaptic cell
the cell producing the signal
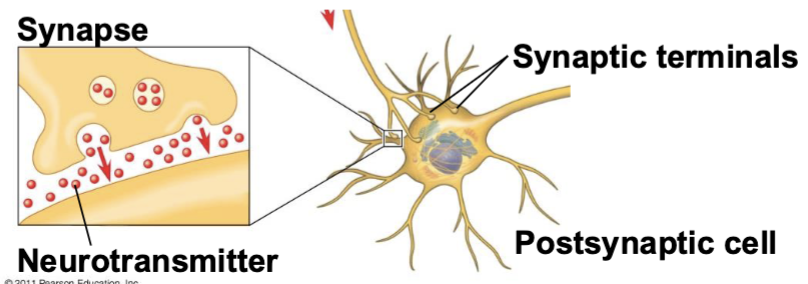
postsynaptic cell
the cell receiving the signal
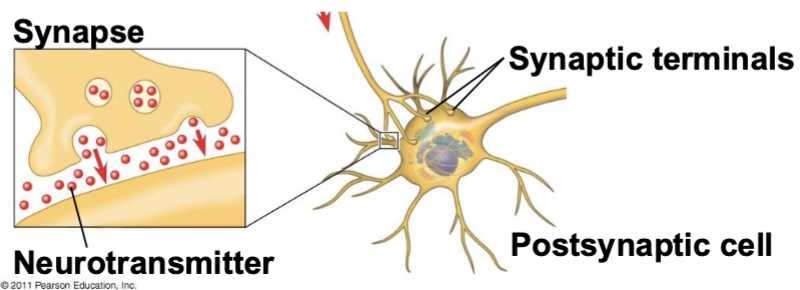
membrane potential
a difference in electrical charge across the cell’s plasma membrane
resting potential
the membrane potential of a neuron not sending signals (at rest, cells normally have excess negative charge on the inside and positive charge on the outside of the cell)
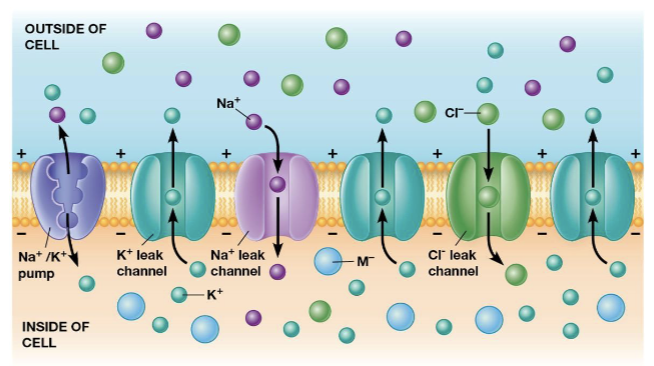
Na+/K+ pump
maintains the voltage by pumping sodium ions out of the cell and potassium ions into the cell
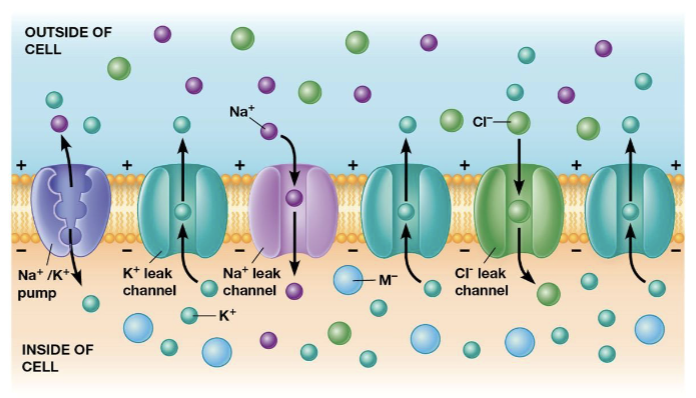
leak channels
allow potassium to diffuse out of the cell, increasing the numbers of anions left behind without counterions (this is how the resting membrane potential is maintained)
action potential
a rapid, temporary electrical signal that travels along the membrane of excitable cells
during, the membrane potential changes from negative to positive and then back again in a very short time
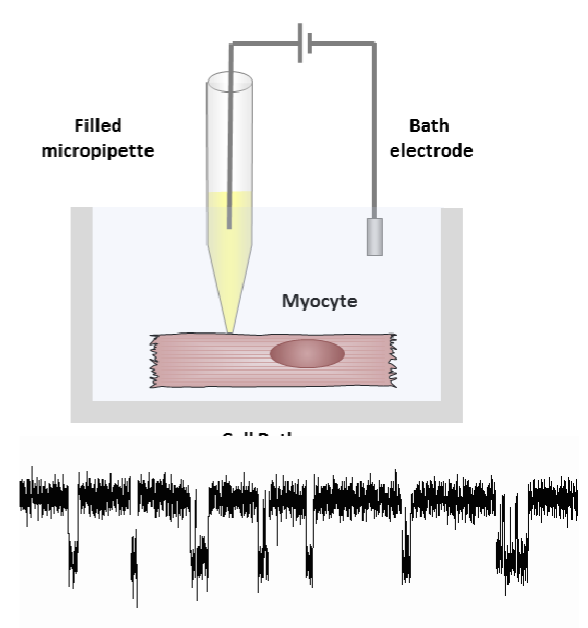
patch clamping
permits the recording of ion currents passing through individual channels, developed by Erwin Neher and Bert Sakmann