quiz #7: murder that never was + DNA rep and repair
1/58
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
59 Terms
what is MMA?
genetic disease where the body cant properly break down certain fats and proteins because of a missing enzyme - methylmalonyl-CoA mutase (MUT)
what does MUT do? what happens when it doesn’t work?
MUT enzyme helps convert methylmalonyl-CoA into succinyl-CoA, which then feeds into the krebs cycle
when this enzyme doesn’t work (MMA), methylmalonyl-CoA builds up and gets converted into methylmalonic acid, which accumulates in the blood and becomes toxic
where is the major site of protein digestion?
small intestine
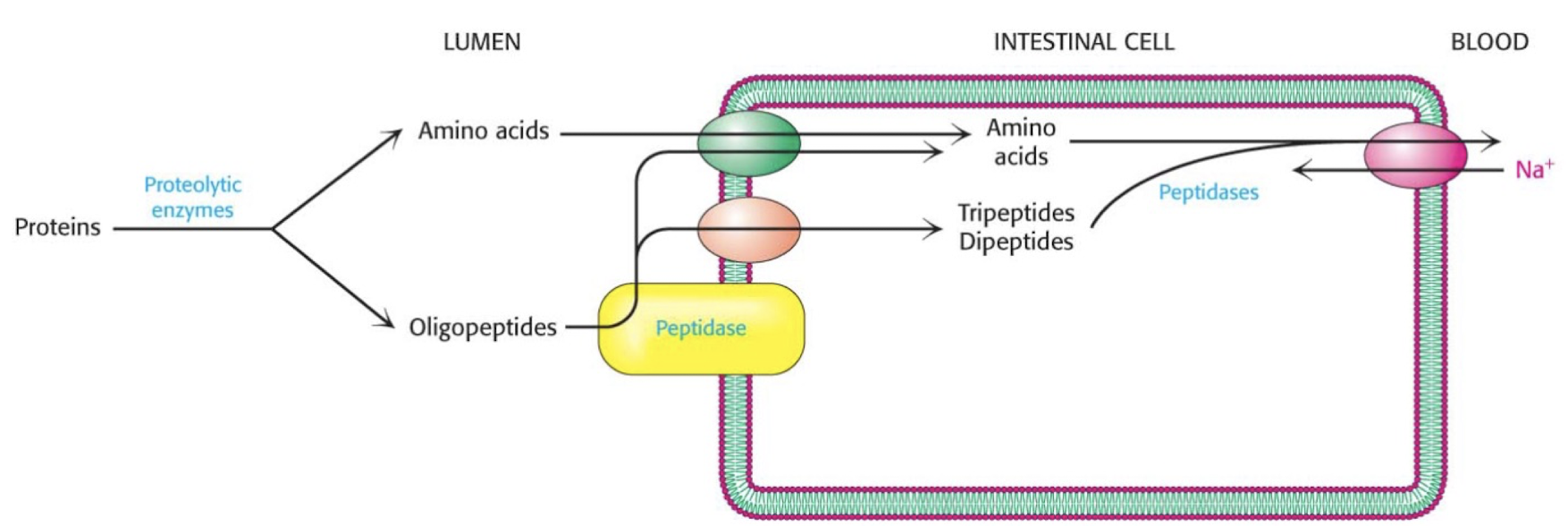
how are proteins digested?
proteins are broken down by proteolytic enzymes in the intestine to amino acids and oligopeptides
AA can be directly absorbed into the intestine, oligopeptides are broken down further by peptidases into amino acids
amino acids are then transported to the blood stream
how are amino acids stored
AA are not stored like fats or carbs, instead the body breaks up the amino group and the carbon skeleton
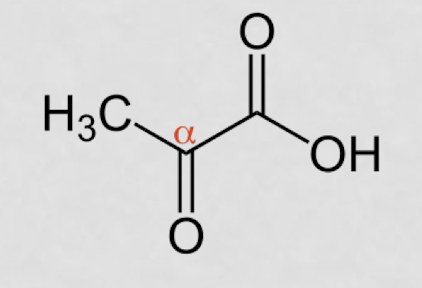
what is an ⍺-ketoacid?
COO- right next to =O
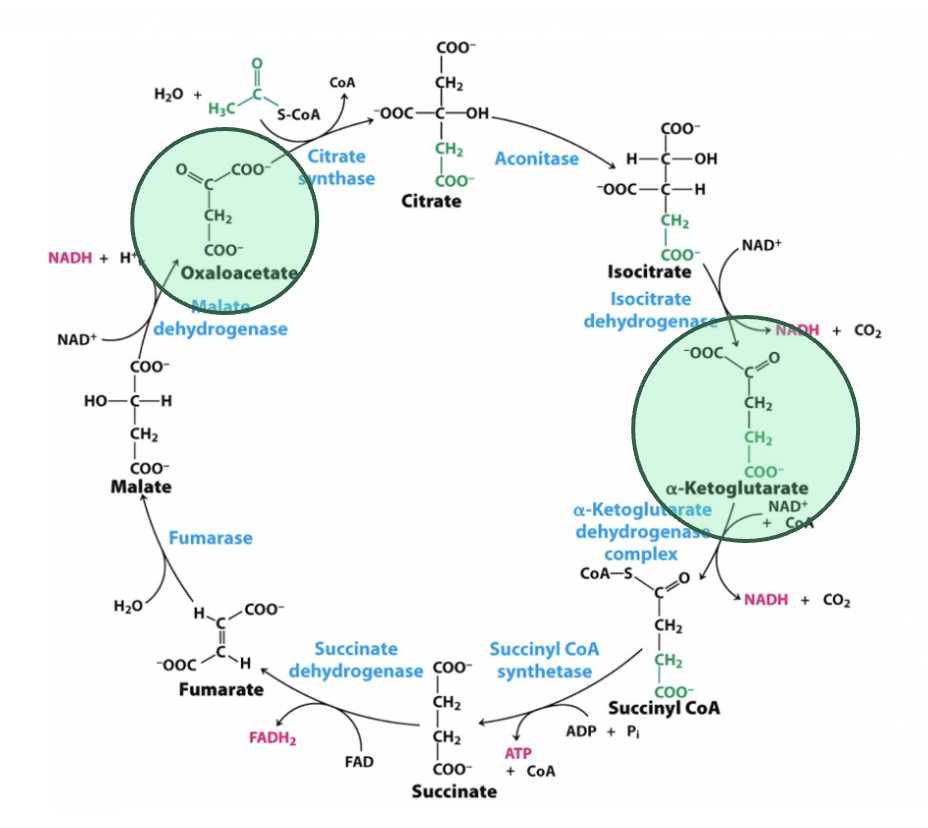
what are the ⍺-ketoacids in TCAC?
oxaloacetate and ⍺-ketoglutarate
what is an ⍺-amino acid?
NH2 group next to COOH
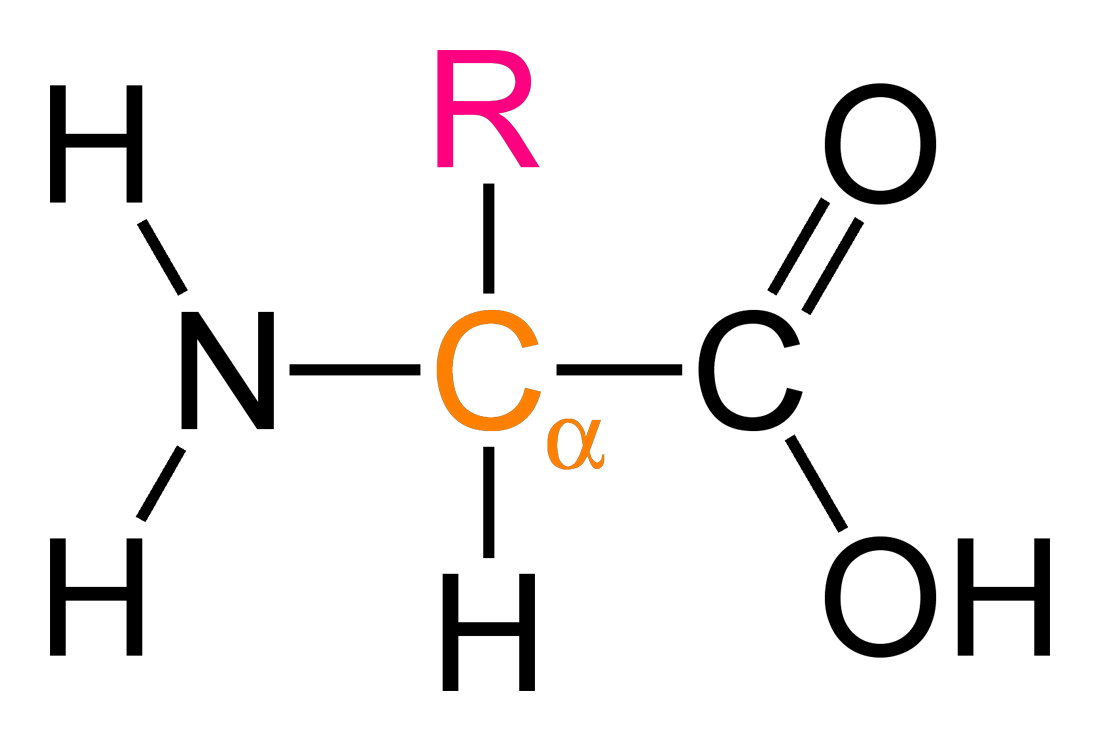
what is oxidative deamination?
the process by which the body removes an amino group from an amino acid to discard the nitrogen
gives the AA to an ⍺-ketoglutarate through an aminotransferasw, creating glutamate and a keto-acid (leftover carbon skeleton)
glutamate is processed by glutamate dehydrogenase where is loses its amino group and produces a free ammonia ionm regenerates ⍺-ketoglutarate so the cycle can start again
energy is captured as NAD+ is reduced to NADH
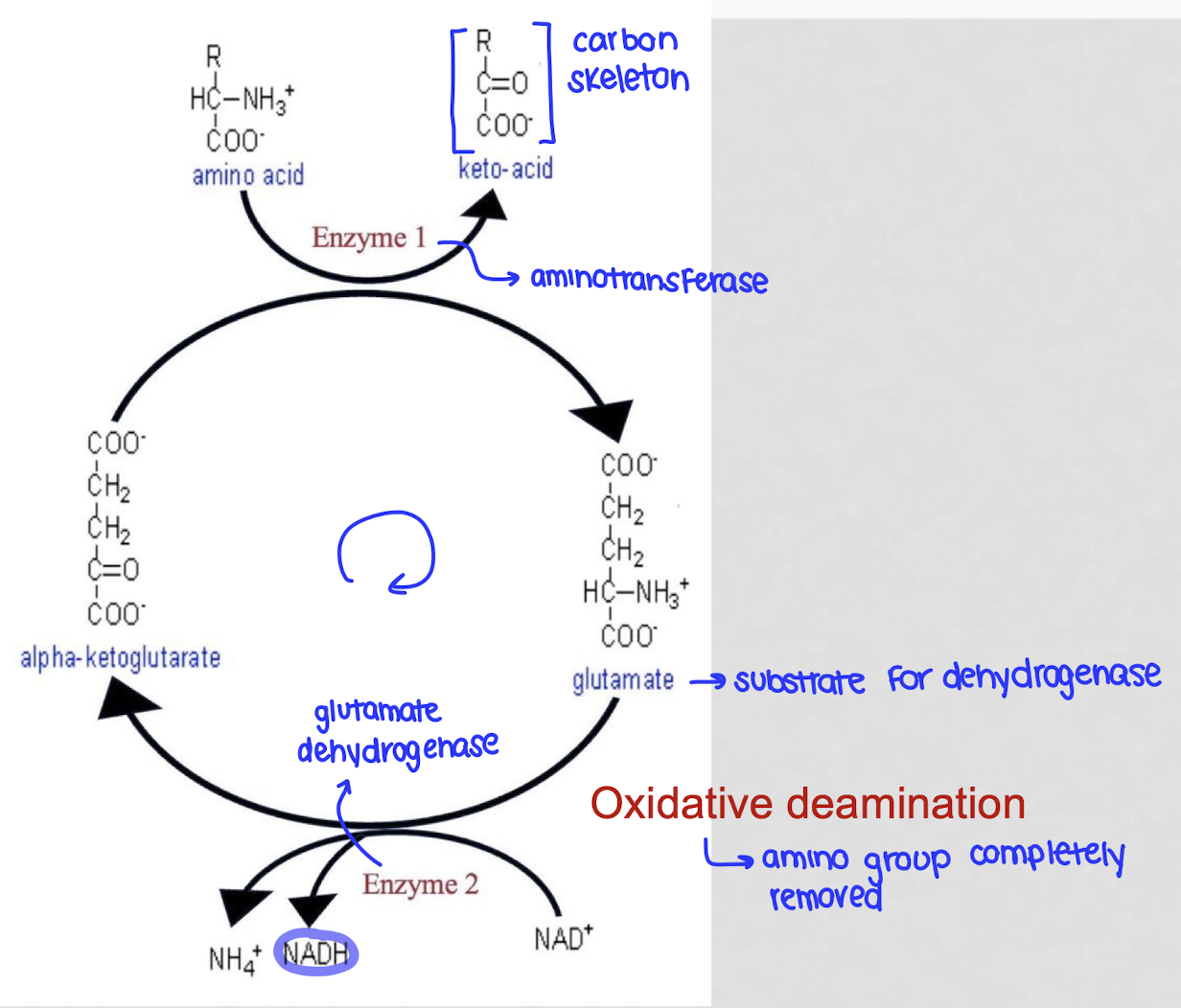
is oxidative deamination catabolic or anabolic?
catabolic - breaks down amino acid
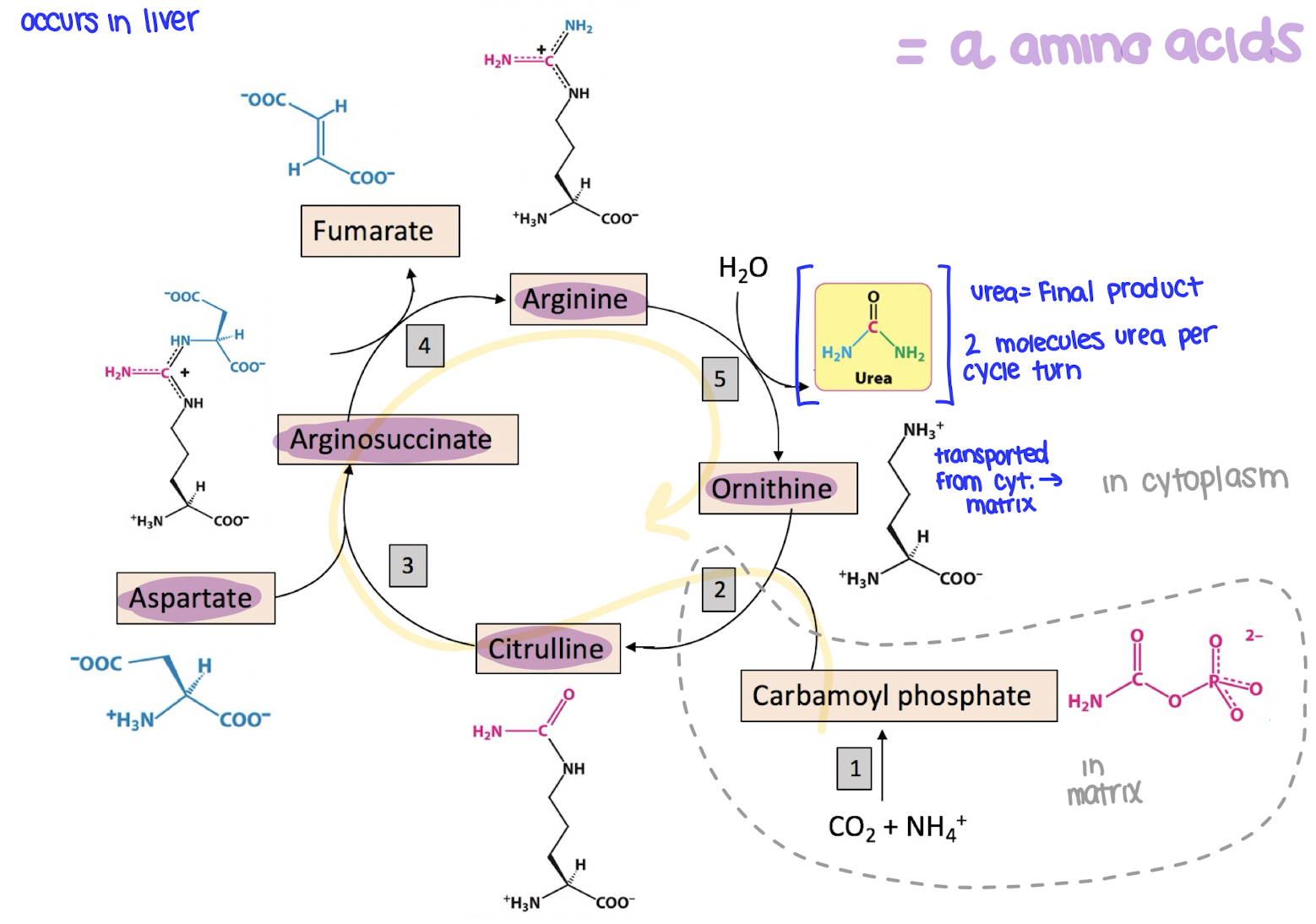
what are the ⍺-amino acids in the urea cycle?
citrulline
aspartate
arginosuccinate
arginine
ornithine
where does the urea cycle occur?
in the liver
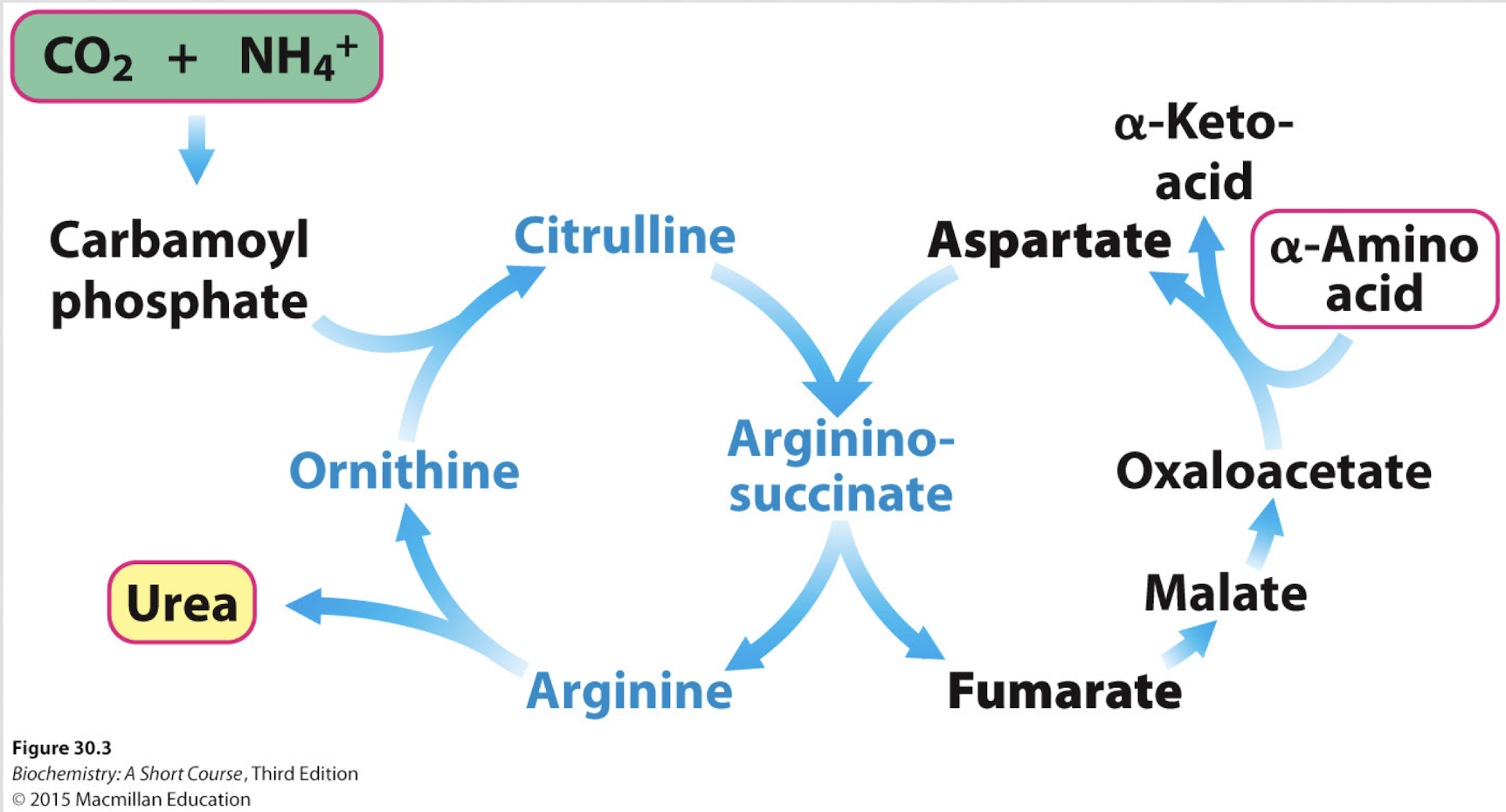
what happens in the urea cycle?
the urea cycle gets ride of NH4+ by turning it into urea, which is then excreted in the urine
ammonia and CO2 combine to form carbomoyl phosphate
carbamoyl phosphate joins ornithine to make citrulline
citrulline combines with aspartate to form arginosuccinate
arginosuccinate is broken into fumarate and arginine
arginine is split into urea and ornithine
urea exits the cycle and is excreted
ornithine is recycled
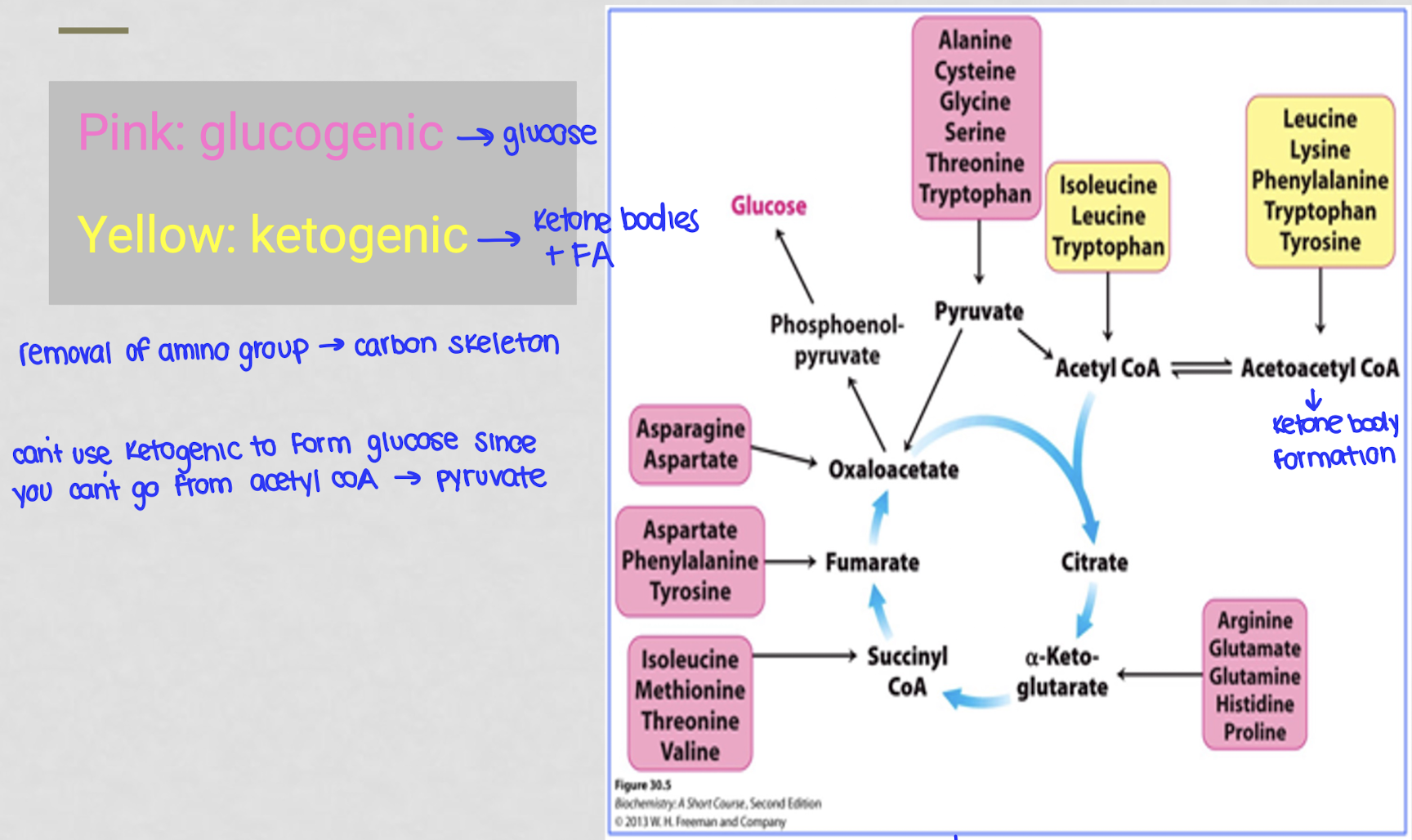
what does it mean to be glucogenic?
anything that can be used to produce glucose
what does it mean to be ketogenic?
anything that can be used to produce ketone bodies + fatty acids
ketogenic carbon skeletons are converted to acetyl coA or acetoacetyl coA which can then be used to synthesize KB and FA
why can’t you use ketogenic things to form glucose?
you can’t go from acetyl-coA to pyurvate
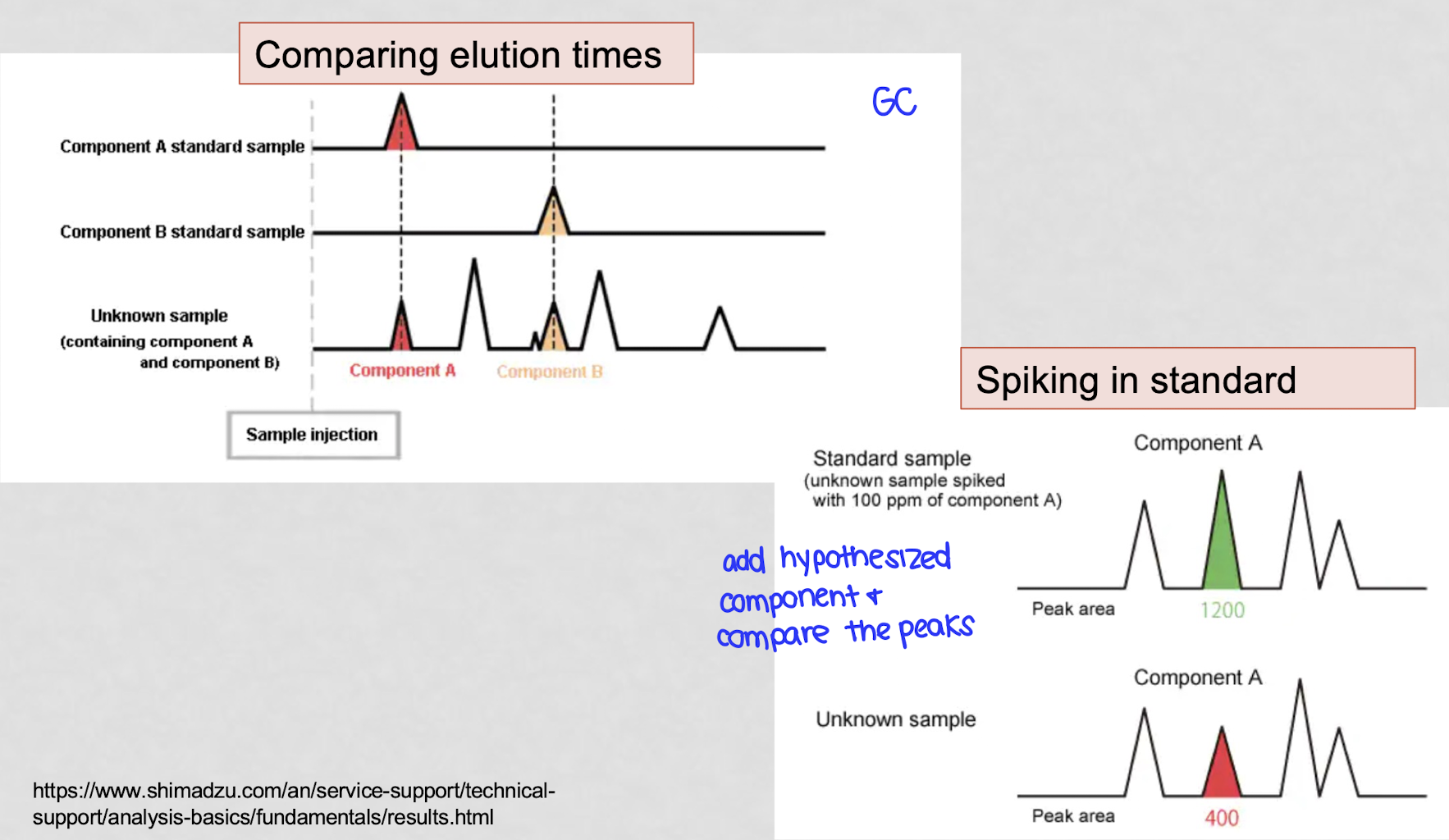
what are 2 things you can do to verify GC peaks?
compare elution times - do each component separately
spiking in standard - add known component and see if a second spike is formed or if that spike grows
where does most amino acid degradation take place?
liver
what are the 2 sources of N in urea?
NH4+ and aspartate
what is the fate of carbon skeletons produced during AA degradation?
can be converted into TCAC intermediates, ketone bodies, or be used to synthesize glucose
what happened in ryan’s case?
Patti was accused of poisoning her baby Ryan with antifreeze and ryan was put into foster care
ryan died from another suspected poisoning after patti fed him a bottle and she was charged with 1st degree murder
ethylene glycol was thought to be found in his flood
in prison, she gave birth to a second baby, DJ
DJ started to have the same symptoms as ryan and was diagnosed with MMA
the case against patti was revisited and tests confirmed that ryan also had mma
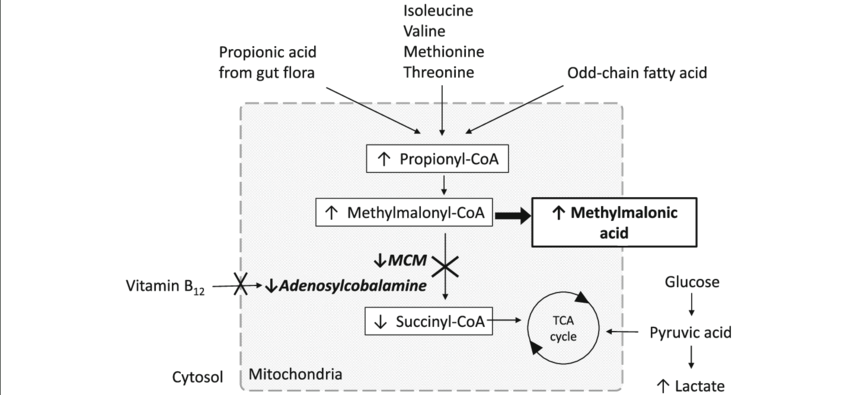
why would a MUT mutation cause elevated levels of propionic acid?
missing MUT → increased levels of methylmalonic coA
propionyl coA and propionic acid which are upstream fo the MUT reaction would increase as equilibrium shifts backwards
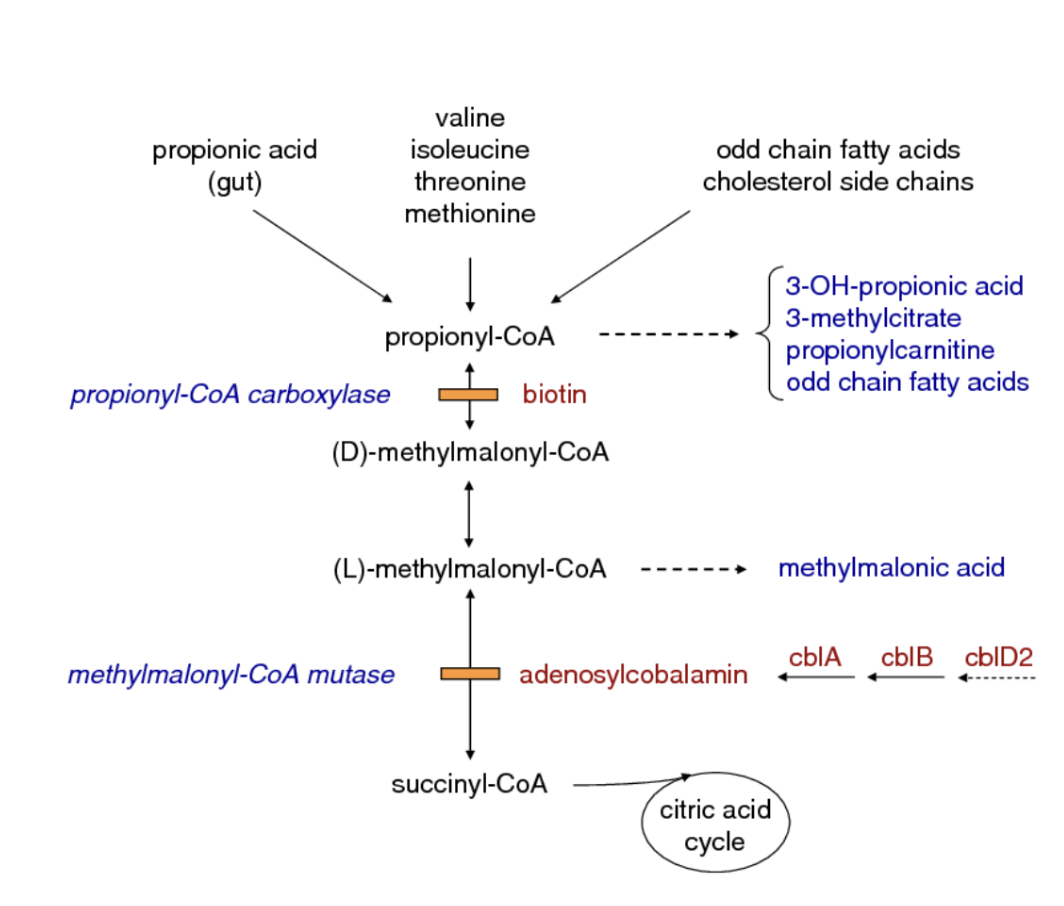
why can a vitamin B12 deficiency mimic a MUT deficiency?
the MUT enzyme requires a coenzyme derived from vitamin B12
without this cofactor, the enzyme cant function even if its normal
this causes a backup of methylmalonic-coA → MM acid build up just like in an actual MUT deficiency
how does a low energy charge affect oxidative deamination?
the reaction will be activated is AMP is high
NADH generated by deamination can then be used by the ETC to produce ATP via oxidative phosphorylation to increase energy charge
why does the step with carbamoyl phosphate synthetase in the urea cycle need to be regulated?
this enzyme uses ammonia which is very toxic if it builds ip
regulated it controls how fast the urea cycle starts, making sure the body only removes nitrogen when needed
what happens to the ⍺-amino acid carbon skeletons after their ⍺-amino acid group is removed?
they get converted to 1 of 7 molecules:
acetyl-coA
acetoacetyl-coA
pyruvate
oxaloacetate
fumarate
succinyl-coA
⍺-ketoglutarate
what is the link between TCAC and the urea cycle?
what were the results of laboratory A in ryans case?
they detected ethylene glycol at 180 mg/L and then at 911 mg/L after he was in foster care
how was propionic acid misidentified as ethylene glycol?
the first lab failed to verify the GC peak and assumed it was EG based solely on the elution time since the GC peak of propionic acid and ethylene glycol are about the same
the second lab spiked in ethylene glycol and saw 2 distinct peaks, meaning the original peak was not ethylene glycol. mass spec confirmed the unknown compound to be propionic acid
LO1: define a transamination reaction and identify alpha keto acids
transamination rxn: when an amino group is transferred from an AA to an alpha-keto acid (no free ammonia released)
alpha keto acid: molecules with C=O next to COOH
LO2: name the enzyme involved in oxidative deamination and how it is regulated in response to energy charge
glutamate dehydrogenase (removes the amino group from glutamate, releasing ammonia and producing alpha ketoglutarate)
low energy charge activates GDH
high energy charge inhibits GDH
LO3: state the purpose of the urea cycle and identify the alpha amino acids of the urea cycle
urea cycle safely removes toxic ammonia (NH4+) from the body by converting it into urea
alpha AA: glutamate, aspartate
LO4: name the 7 molecules that are produced from the carbon skeletons of amino acids after deamination
pyruvate (G)
acetyl-coA (K)
acetoacetyl-coA (K)
alpha-ketoglutarate (G)
succinyl-coA (G)
fumarate (G)
oxaloacetate (G)
LO5: differentiate between ketogenic and glucogenic amino acids
GG: after breakdown they make molecules that can be turned into glucose and then feed into the krebs cycle
KG: after breakdown they make molecules that can be turned into ketone bodies or FA
LO6: state the function of the enzyme deficient in MMA and how that relates to lipid and AA metabolism
enzyme - methylmalonyl-coA mutase (MUT) which converted methylmalonyl-coA into succinyl coA which then enters the krebs cycle
FA and AA are broken down into MM-coA, which needs MUT to be process
without this MM-coA builds up and energy production from this is blocked
what are the purines and pyrimidines?
purines: AG (pure as gold) - 2 rings
pyrimidines: CUT (pyraminds cut) - 1 rings
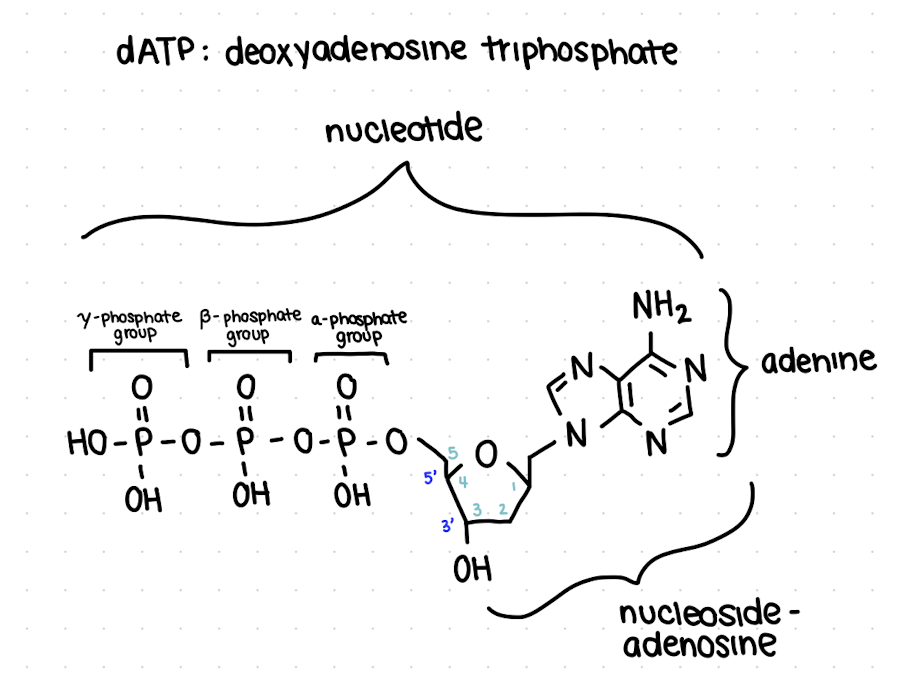
how are nucleotides named?
[deoxy]
name of base
# of phosphates
phosphate
![<ol><li><p>[deoxy]</p></li><li><p>name of base</p></li><li><p># of phosphates</p></li><li><p>phosphate</p></li></ol><p></p>](https://knowt-user-attachments.s3.amazonaws.com/97050a3f-4531-4493-993b-dd68ccbc8d26.png)
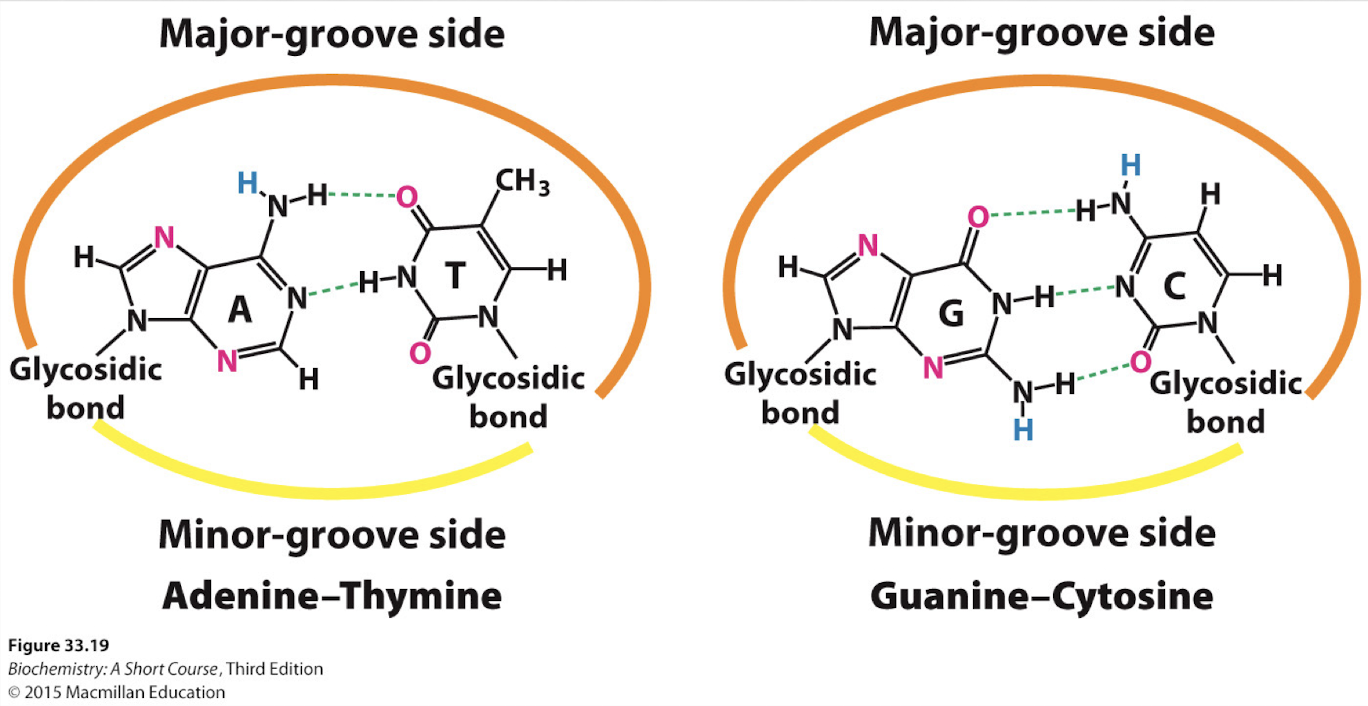
what are the base pairs?
A : T/U - 2H
C : G -3H
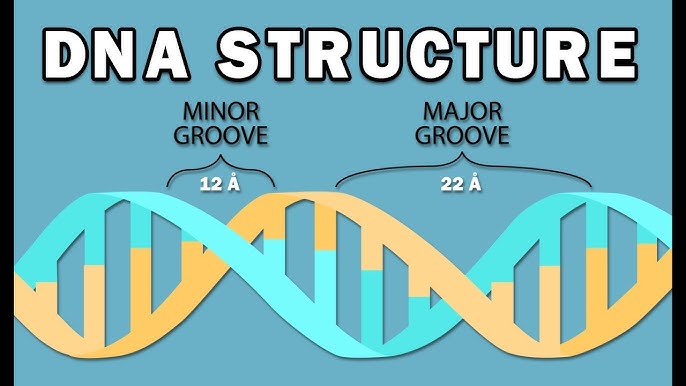
what are the major and minor grooves in DNA?
major groove: wider and deeper gap
minor groove: narrower and smaller gap
proteins often read the DNA by binding to the major groove
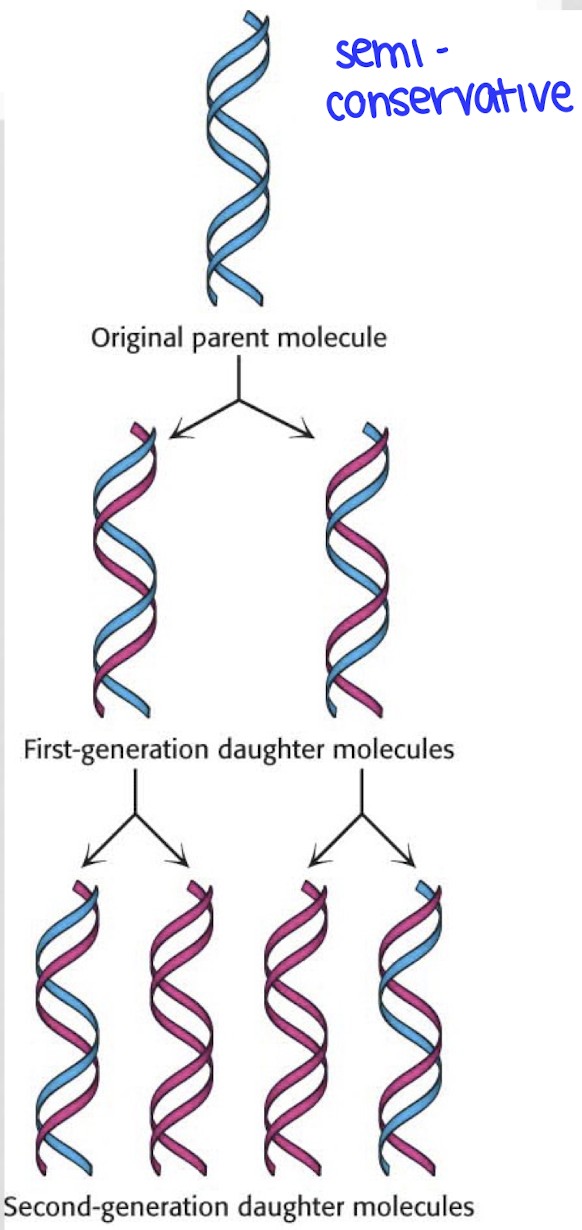
what does it mean that DNA is semiconservative?
the two complementary strands of DNA come apart and each serve as a template strand for a new DNA strand
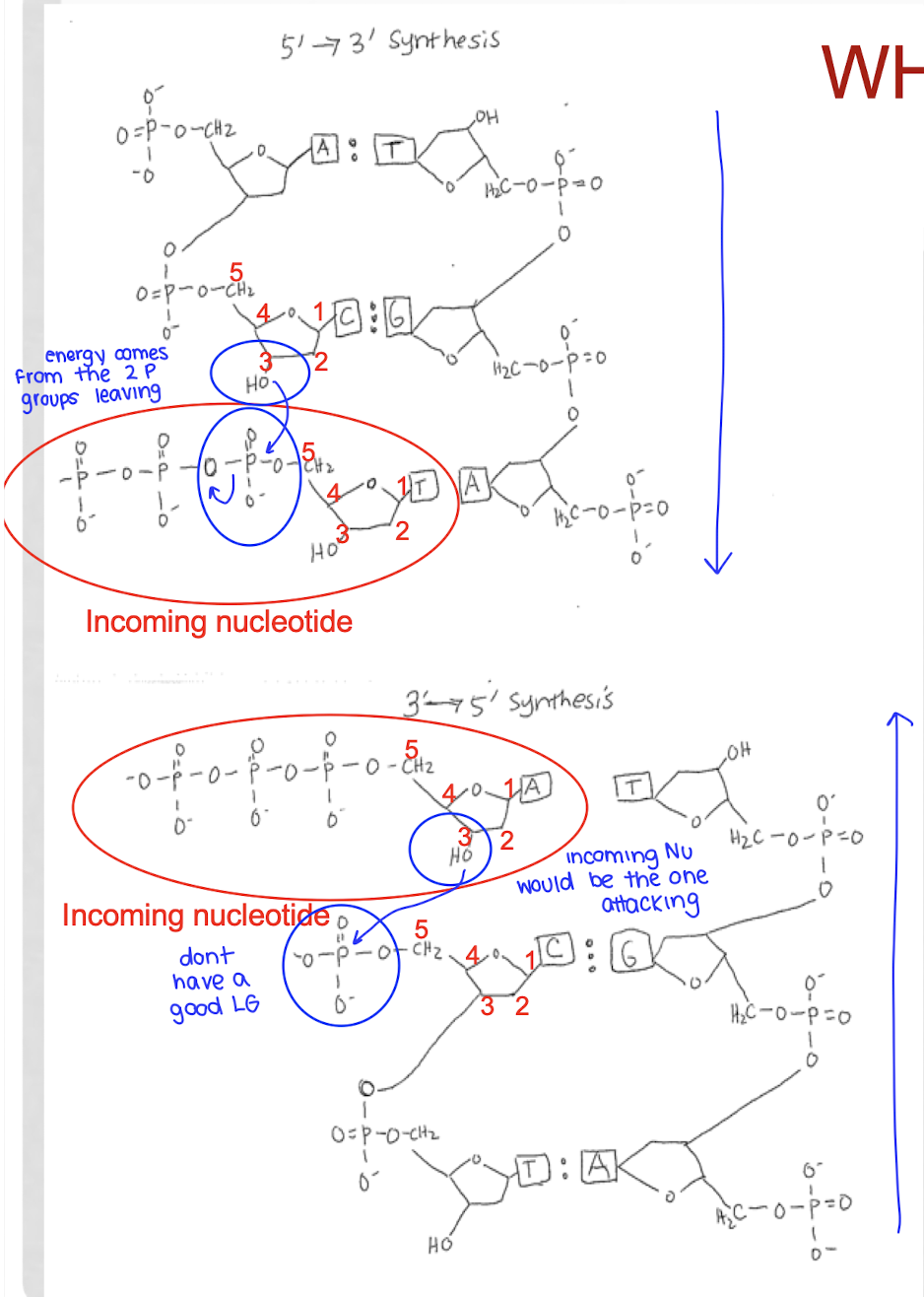
how is a new nucleotide added onto a DNA strand?
the 3’ OH of the last nucleotide of the DNA strand attacks the alpha phosphate of the first phosphate group of the incoming nucleotide, kicking out a diphosphate → phosphodiester bond
catalyzed by DNA polymerase
ribose is the nucleophile, a-P is the electrophile
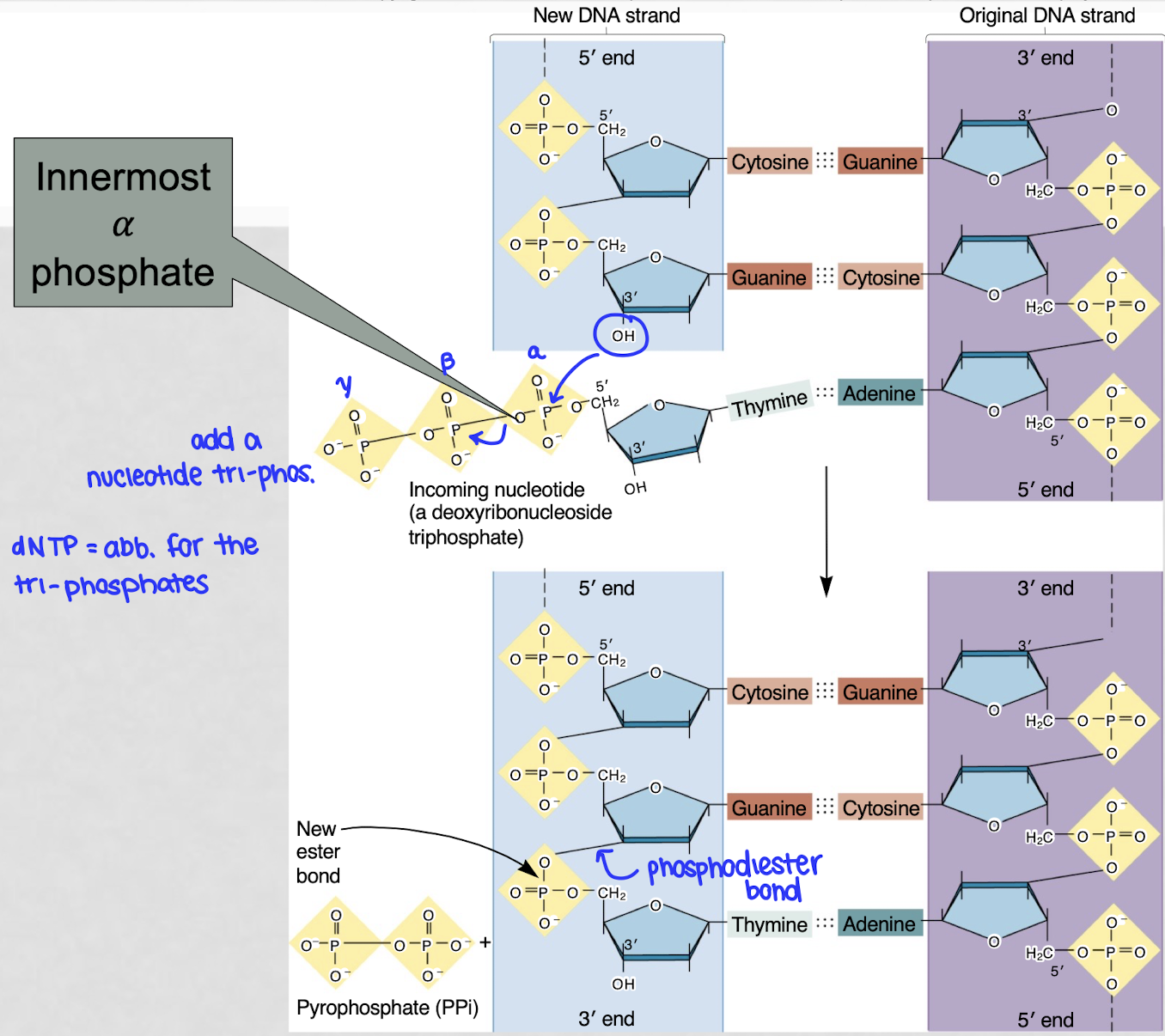
which end does DNA grow?
adds nucleotides onto the 3’ end
what is a nucleoside?
base + sugar (no phosphate)
where do you start numbering from on the ribose sugar?
start at the C to the right of O - OH should be at 3 (3’ OH)
what 2 types of non-covalent interactions contribute to the formation and stabilization of the DNA double helix?
VDW occurs between the stacked bases of one strand
HB occurs between the bases connecting two strands
what are the substrates of DNA polymerase?
primer, parent strand (template), a deoxynucleotidetriphosphate (dNTP - incoming nucleotide)
what atoms are directly involved in the reaction with DNA polymerase?
the 3’ OH of the last added nucleotide attacks the alpha phosphorus atom of the incoming nucleotide
what is lost from the incoming nucleotide by DNA polymerase?
2 PPi
what is the role of Mg2+ with DNA polymerase?
2 ions required
one stabilizes the O- of the 3’ carbon on the primer strand, increasing its nucleophilicity leading to the attack
the other stabilizes the negative charges on the PPi leave group
(One of the magnesium ions is used to stabilize the negative charge of the 3' oxygen anion of the last nucleotide of the primer strand in order to increase its ability to attack as a nucleophile. The other magnesium ion is used to stabilize the negative charges on the β and γ phosphate groups that leave the molecule.)
why can DNA replication not occur in the 5’ → 3’ direction?
because then the reaction couldn’t be favored with the release of PPi, which is very favorable
the oxygen with the w P is a good LG to allow the bond to form between the 3’ O and the a-P of the incoming nucleotide
if it tried to proceed in the 3’ → 5’ direction?
the 3’OH of the incoming nucleotide would have to attack the P group of the last added nucleotide
that is not possible as there is no good LG
if replication were to occur in the 3' to 5' direction, this energy would come from the 5' phosphate group already incorporated into the nucleotide, which is a less stable leaving group and thus less energetically favorable
where are the hydrogen bond donors and acceptors in base pairing?
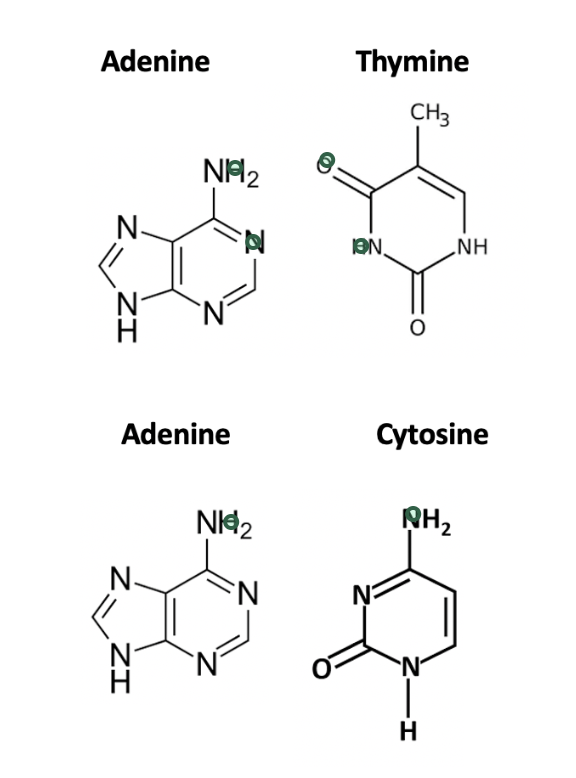
what 2 reactions are necessary to convert C → T?
(1) a deamination reaction converts the amine group at the top to an keto group
(2) a methylation reaction adds a methyl group adjacent to the keto group
what are the sites in a DNA polymerase enzyme?
polymerase catalytic site (palm): where new nucleotides (dNTPs) are added to the growing DNA strand by a phosphodiester bond
3’ exonuclease site: where proofreading happens
removes incorrect bases in the 3’ → 5’ direction
are mistakes during DNA replication common or rare?
extremely rare1
what are the reasons why fidelity in DNA replication is high?
instability of mismatched pairs
complementary BP have a much higher stability than mismatched pairs
configuration of the DNA polymerase active site
DNA p is unlikely to catalyze bond formation between mismatched pairs
proofreading function of DNA polymerase
DNA p can identify a mismatched nucleotide and remove it from the daughter strand
what causes DNA polymerase to pause or slow down during the process of DNA synthesis?
when a wrong NT is incorporated during DNA synthesis, the enzyme stalls due to the structural instability by the incorrect base pairing
since the proper number of hydrogen bonds is required for a pairing to be stable, an error in this will cause the base pairing to be highly unstable, which DNA polymerase can detect due to the movement
the pause provides time for the incorrectly synthesized strand to be moved to the exonuclease active site of the enzyme, where wrong NT are removed
once the NT is removed, the strand is moved back to the polymerase active site and synthesis continues
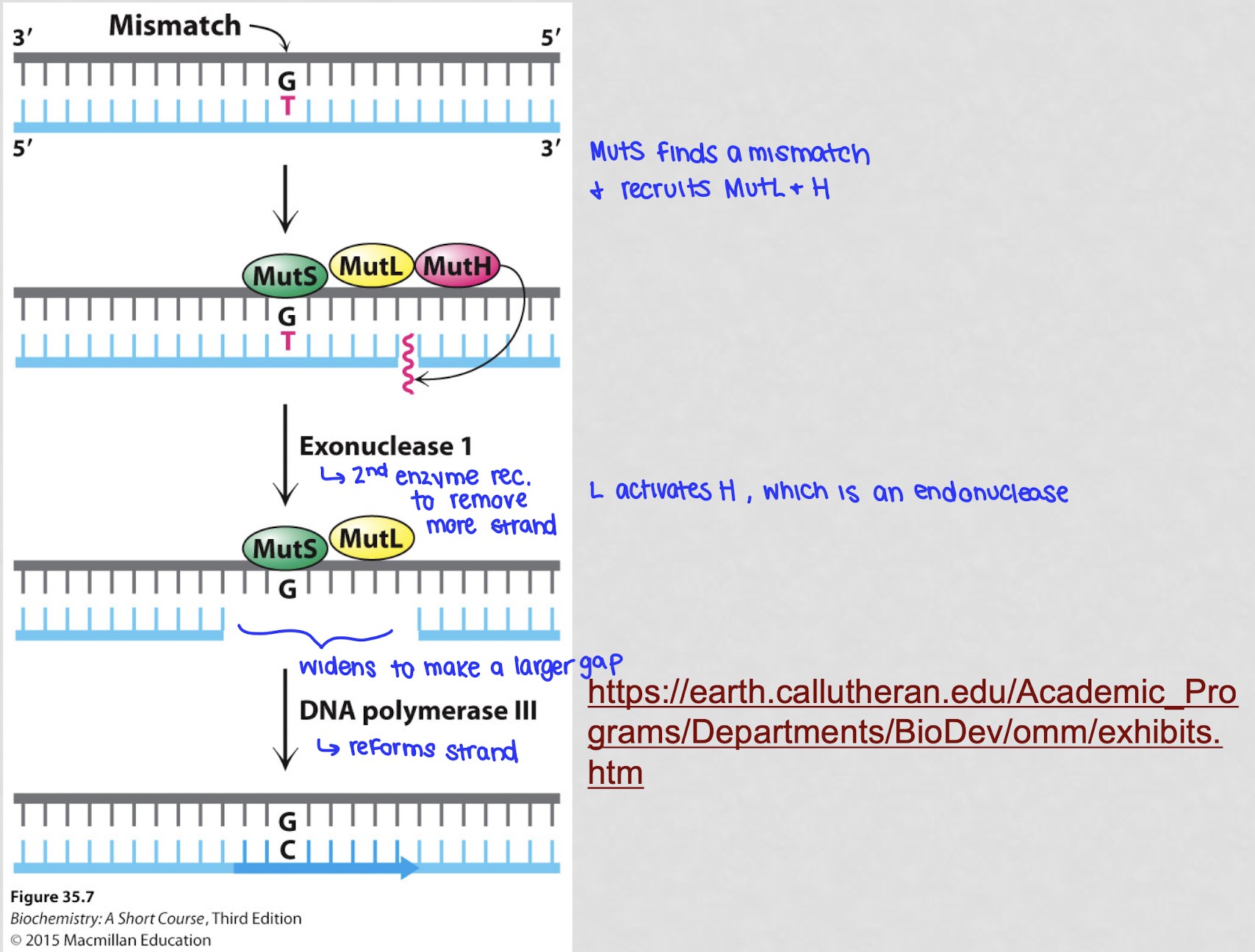
what happens during the process of mismatch repair?
if the 3’ → 5’ exonuclease doesn’t catch it right away, MM repair steps in
the MutS protein complex finds the mismatch by recognizing the distortion caused by the wrong BP
MutL binds are helps coordinate the repair by finding which strand is new (not methylated)
MutH cuts out a section of the wrong DNA strand, including the mismatch
DNA polymerase fills the gap with the correct NT
MutS recognizes the incorrect BP and recruits MutL and MutH. MutL activates MutH which is an endonuclease that cleaves the newly synthesized DNA strand close to the mismatch
describe the MutS monomer domains?
the clamp domains of each monomer initially recognize the mismatched DNA and have a positive charge on their surface, allowing for electrostatic attractions to form with the negatively charged DNA backbone
these interactions are sequence independent
the DNA molecule can now make contact with the mismatch-recognitition monomer of MutS through its bases and the non-mismatch binding monomer of MutS through its backbone
compare the 2 MutS monomers?
mismatch recognition monomer:
binds the major groove
base-specific interactions
uses the mismatch-binding domain
non-mismatch binding monomer:
binds the minor groove
electrostatic interactions
uses the clamp domain