Cell and molecular Biology test number 3
1/21
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
22 Terms
Protein post-transitional modifications (PTM)
Amino acid modification
phosphorylation (PO4³-,usually used for signal transduction)
cofactor binding (enzymatic activity)
Glycosylation (addition of oligosaccharides)
Acetylation (-CO-CH3; ON LYSINES OF HISTONES)
Lipidation
Cleavage of peptide bonds and protein processing
Ubiquitination (for signal transduction, or proteasomal degradation)
SUMOylation
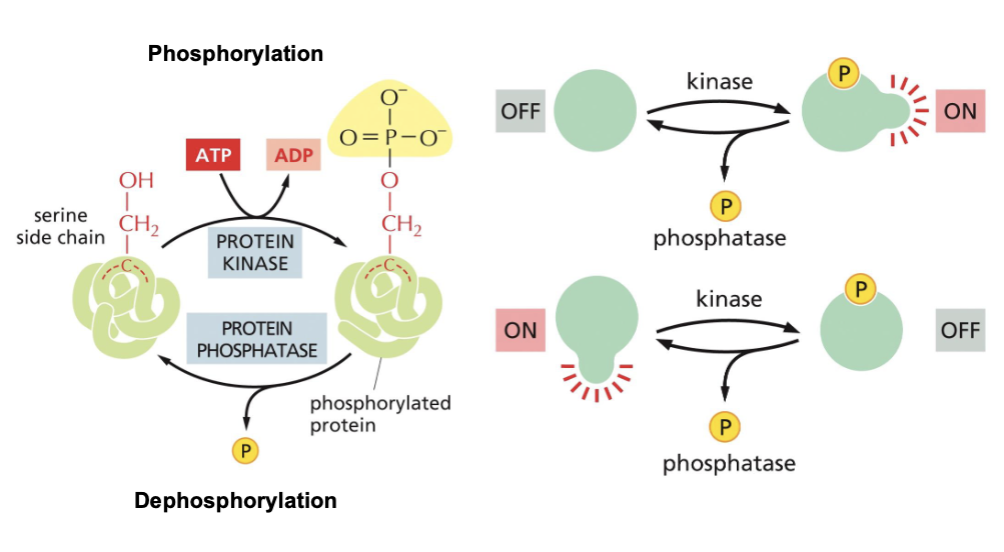
Proteins are modified by phosphorylation
The three most common phosphorylated amino acids are serine, threonine, and tyrosine. A non-polar amino acid can become polar via addition of a PO4³- group.
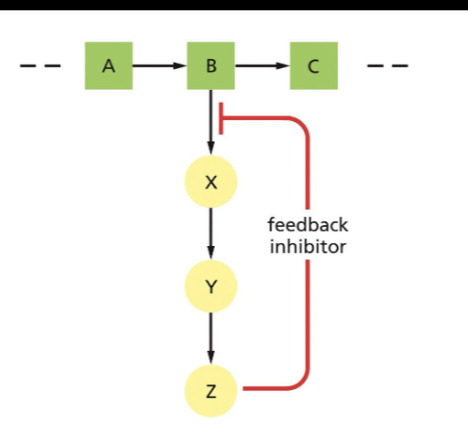
Feedback inhibition and allosteric binding
Allosteric regulation is the regulation of a protein by the binding of an effector molecule at a site other than the active site: Allosteric activation or allosteric inhibition.
In a negative feedback inhibition, an enzyme of allosteric inhibition, a product of the metabolic pathway, inhibits one of the enzymes that produces an intermediate metabolite of the pathway.
-This example shows the feedback inhibitor is Z, which is inhibiting the enzyme responsible for catalyzing the reaction of converting B into X.
They are in LacI→ Allosteric inhibition and TrpR, where this one is Allosteric activation.
Feedback inhibition of Anthranilate synthesis
Binding of tryptophan to an allosteric site

Aspartate-family biosynthetic pathway
Includes five photogenic amino acids in the metabolic pathway, which are highlighted with an asterisk next to them.
Aspartate Kinase (AK) is inhibited by a different metabolic intermediate. Two of these AK are bifunctional: they are also homoserine dehydrogenases (HDH).
AKs are not present in animals.
Allosteric Activators increase protein activity
Why would you want to increase enzyme activity when bound to ADP?
-ADP indicates low ATP concentration in the cell; therefore, glucose should be catabolized.
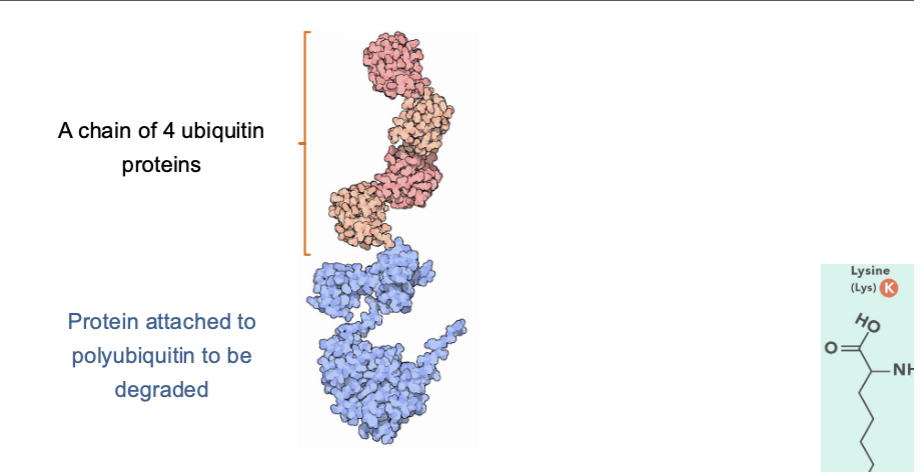
Ubiquitin Protein
The ubiquitin protein is added to other proteins to tag them for degradation. And to regulate the function of other proteins.
Ubiquitin chains are added to specific lysine amino acids in the proteins and there are 4 for chain.
The chains that tag a protein for degradation are formed by adding new ubiquitin molecules to lysine 48 of ubiquitin.
Ubiquitination
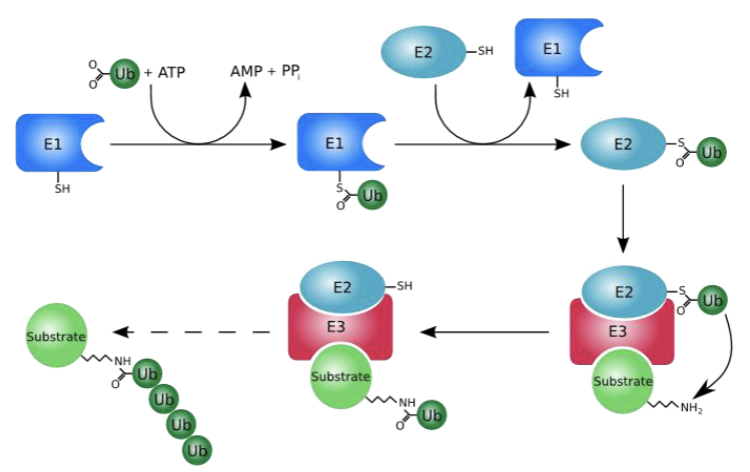
This is a protein modification that happens with the aid of 3 enzymes and is post-translational
The process of attaching ubiquitin to a target protein is a multi-step enzymatic cascade that requires energy.
-E1=ubiquitin protein-activating E1 enzyme, then transfers to E2=Ubiquitin conjugating enzymes which is conjugation step; THEN —> E3=ubiquitin ligases, which recognize the protein substrates. And transfers the ubiquitin from the E2 enzyme to the target protein, forming a covalent bond.
Ub=ubiquitin
It can act as a switch, turning cellular signaling pathways on or off by changing a protein’s function or localization.
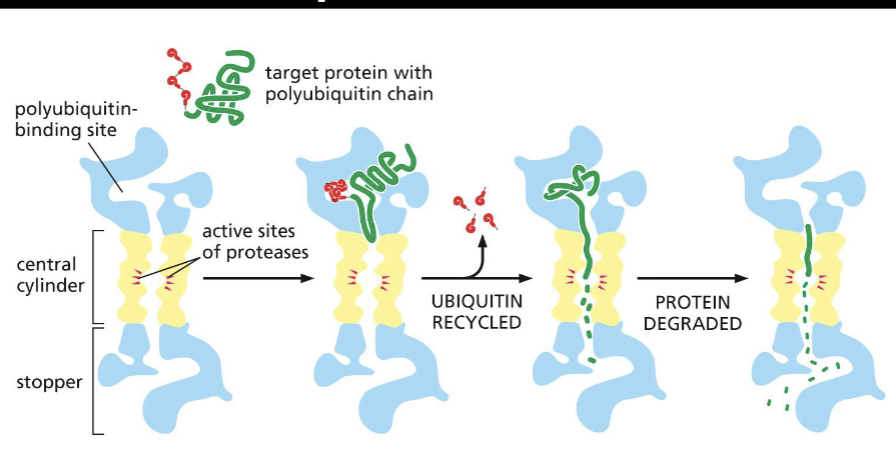
Proteasome
The proteasome is a multi-protein complex that degrades proteins. This has ubiquitination proteins in the proteasome.
-It degrades misfiled and damaged proteins, or degrades proteins that are no longer needed by the cell (recycling their amino acids). They are present in the cytosol and nuclei of eukaryotic cells.
Ubiquitinated proteins
These proteins are degraded by the proteasome
ubiquity molecules are cleaved off before protein destruction by the proteasome so they can be reused.
Proteins are rapidly degraded inside the proteasome into small 3-25 amino acid polypeptides.
How can proteins localize to different cell compartments?
This is by protein targeting or sorting
Proteins cannot traverse the hydrophobic membranes on their own.
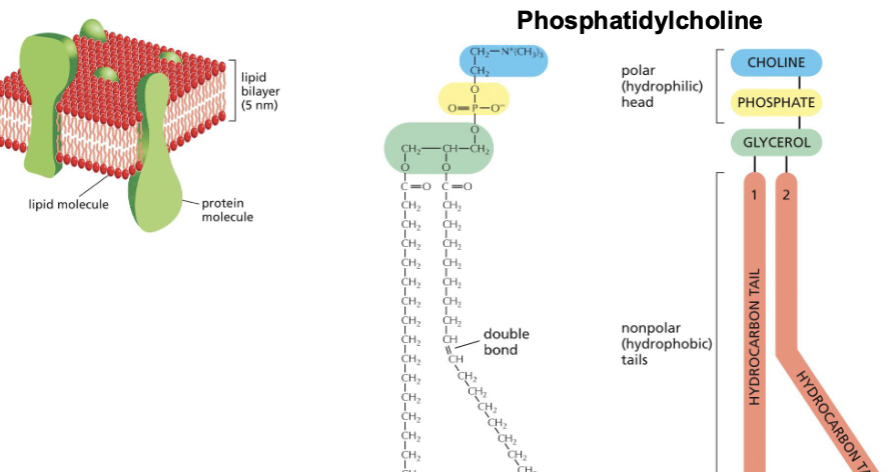
Mmembranes are mostly composed of lipids and proteins
What are three other parts of a phospholipid?—> choline, phosphate, and glycerol.
Phosphatidylcholine is one of the most common phospholipids present in the membranes of animals and plants.
hydrocarbon tails are fatty acids, attached to glycerol by their carboxyl (cool) group.
Protein associations with lipid bilayer membranes
Integral membrane proteins are membrane proteins permanently attached to the membrane.
Its hydrophobic inside and outside the cell is hydrophilic.
They transport molecules send signals and help cells adhere to each other.

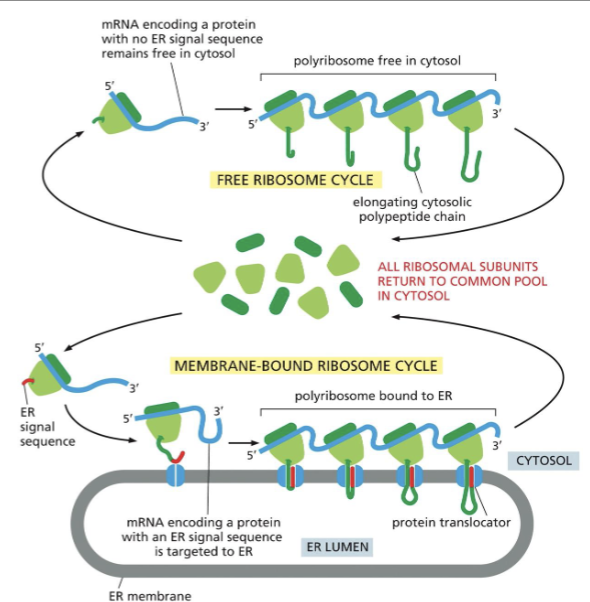
Protein transport through the cell.
all proteins are synthesized in ribosomes in the cytosol.(some free floating, and some attached to the ER).
1) Nuclear pores: proteins are transported FOLDED.
2) Across membranes: Proteins need to be transported across the membranes, and refolded after transport, this is done by protein translators. Same mechanism as the one usually observed in bacteria.
3) By Vesicles: Proteins are transported FOLDED.
LUMEN
Its the inside space defined by a membrane in some organelles.
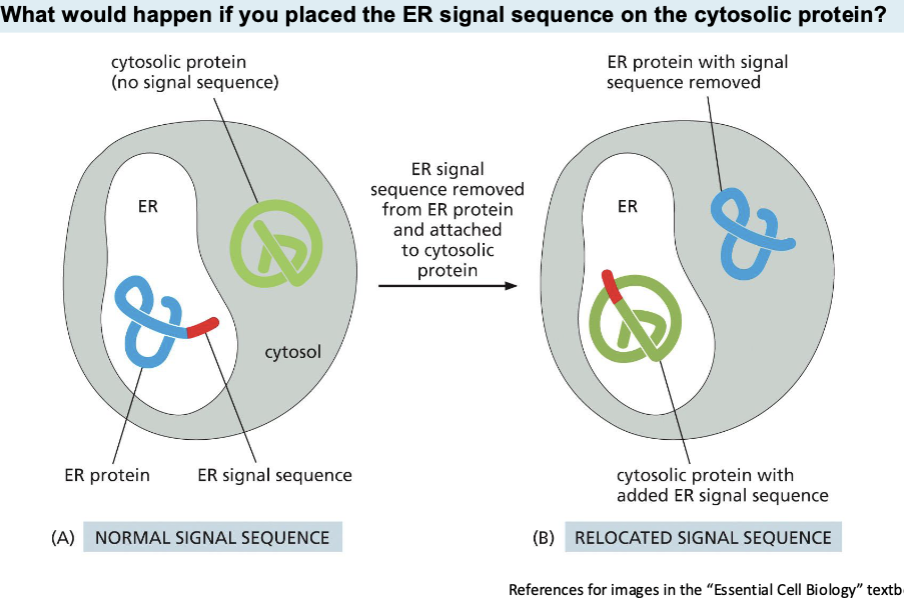
Signal Sequences
On their own direct proteins to the right cell compartments
what would happen if you placed the er signal sequence on the cytosolic protein?—> The protein will be targeted for entry into the ER, where it will be fully synthesized and processed.
Outer membrane and ER: One and only 1) Nuclear Pores
There are 2 concentric membranes (nuclear envelope) surrounding the nucleus. The outer nuclear membrane is continuous wit the ER membrane.
Nuclear pores facilitate transport between the cytosol and nucleus.
Smaller molecules diffuse through the nuclear pores. RNA and ribosomal proteins move from the nucleus to cytosol, while proteins move into the nucleus.
Each pore can be involved in up to 1,000 translocations per second.
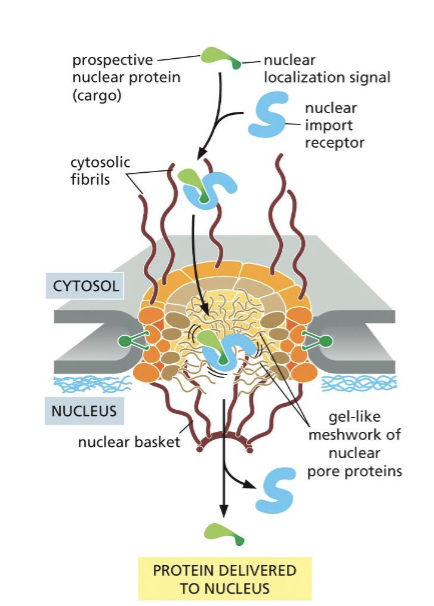
Delivery of proteins inside the nucleus
Proteins imported into the nucleus carry a nuclear localization signal (NLS) recognized by the nuclear import receptor (importins), which transports the protein between the cytosol and the nucleus.
Exportins are involved in the passage of proteins from the nucleus to the cytosol and bind to nuclear export signals (NES).
The center of the nuclear pore complex is filled with proteins that loosely bind to one another in a gel-like structure (FG Nup).
the importing bound to their protein cargo bind to The-Gly nucleoporins of the nuclear pore complex (NPC).
Ran-GTP cycle
Ran is a small GTPase that can either bind GTP or GDP.
-Ran-GTP is primarily located in the nucleus, while Ran-GDP is located in the cytosol.
-Ran-GAP (GTPase activating protein) which facilitates GTP hydrolysis, is only in the cytosol.
-Ran-GEF (Guanine nucleotide exchange factor) which exchanges GDP for GTP in Ran is only in the nucleus.
-The cycle is involved in import and export into the nucleus.
In the nucleus, Ran-GTP binds to the importin (receptor), releasing the cargo into the nucleus.
The importin-Ran-GTP complex is transported to the cytosol, where GTP is hydrolyzed.
Ran-GDP dissociates from the importin, and now the importin is able to bind to a new protein cargo.
GTP hydrolysis drives nuclear transport.
Protein export from the nucleus
A tripartite complex of Ran-GTP, exporting, and a protein with an NES (nuclear export signal) are exported the nucleus through the NPC (nuclear pore complex).
Once GTP is hydrolyzed to GDP (Ran is a GTPase), the cargo protein is released into the cytosol.
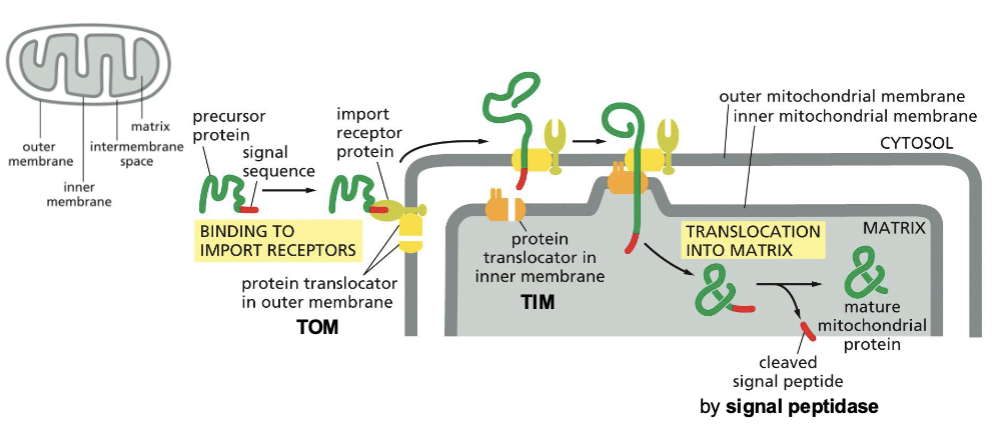
(2) Across membranes:Translocation into mitochondria
After protein synthesis in cytosolic ribosomes, proteins are imported Ito mitochondria and chloroplasts. Proteins carry a mitochondrial or chloroplast signal sequence in their N Terminus, which is cleaved after import.
N terminus is the ned of a protein chain with a free amino acid group (-NH2) and comes first in protein synthesis and can contain signals for protein targeting to specific locations
C terminus is the end with free carboxyl group of (COOH). Comes last and helps terminate protein synthesis and sorting or be used to anchor proteins to membranes.
Proteins are usually transported unfolded.
Mitochondria protein translocators:TOM AND TIM, translocase for the outer and inner membrane of the mitochondria, respectively.
Translocation through both membranes occurs simultaneously.
Translation and translocation into the ER are usually interlinked
Proteins without an ER signal sequence are synthesized in cytosolic ribosomes.
Proteins ER signal sequence and the SRP( Signal recognition Particle) direct the ribosome towards the ER (cotranslocational translocation)