Lecture 19: Non-Coding RNAs
1/19
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
20 Terms
Long noncoding RNA (lncRNA)
Long RNA molecules that tend to be very large (>200 nucleotides) that do not code for proteins but function in regulating gene expression
Ex. Xist and Tsix
Some can serve as a “sink” that absorbs small non-coding RNAs
Can be transcribed from enhancers, intergenic regions, exons, and introns
Many contain a 5’ cap and 3’ poly(A) tail, similar to mRNA — but unlike mRNA, lncRNA is not transcribed
Most abundant in the brain and CNS
List the types of RNA that are present in both prokaryotes and eukaryotes.
Messenger RNA (mRNA)
Ribosomal RNA (rRNA)
Transfer RNA (tRNA)
List the types of RNA that are present only in eukaryotes.
Pre-messenger RNA (pre-mRNA)
MicroRNA (miRNA)
Small interfering RNA (siRNA)
Piwi-interacting RNA (piRNA)
List the types of RNA that are present only in prokaryotes.
CRISPR RNA (crRNA)
Xist and Tsix are examples of…
Long non-coding RNA (lncRNA)
Long non-coding RNA (lncRNA) is approximately…
>200 nucleotides long
Piwi-interacting RNA (piRNA) is approximately…
26-31 nucleotides long
Piwi-interacting RNA (piRNA)
The largest class of small non-coding RNA molecules expressed in animal cells
Single-stranded RNA molecules that are 26-31 nucleotides long and are involved in the post-transcriptional silencing of transposable elements
RNA-mediated adaptive immunity against transposition
piRNA is transcribed and binds to the Piwi protein to form a complex
Piwi protein guides piRNA to its complementary sequence, which is either within the transposon or the transposase → triggers cleavage and degradation of transposon and transposase RNA, preventing transposition from happening
Dysgenesis
Abnormal organ development during embryonic growth and development
Ex. Hybrid dysgenesis in Drosophila is caused by the transposition of P-elements
P+ indicates presence of P-element
P- indicates absence of P-element
P-elements produce piRNAs that inhibit transposition of P-elements, and piRNAs are found only in the egg (maternal gamete)
Sperm do not transfer piRNA during fertilization
If a P- male and P+ female cross, the paternal gametes lack P-elements and the maternal gametes contain P-elements → maternal P-elements produce piRNAs so P-elements are not transposed → no hybrid dysgenesis and development is normal, producing fertile offspring
If a P+ male and P- female cross, the paternal gametes contain P-elements but the maternal gametes lack P-elements → no piRNAs are produced, so P-elements in the paternal chromosomes undergo a burst of transposition → resulting in hybrid dysgenesis, mutations, chromosomal aberrations, and sterile offspring
P-elements
Segments of DNA in Drosophila (and other organisms) that can move within the genome
P+ indicates presence of P-elements
P- indicates absence of P-elements
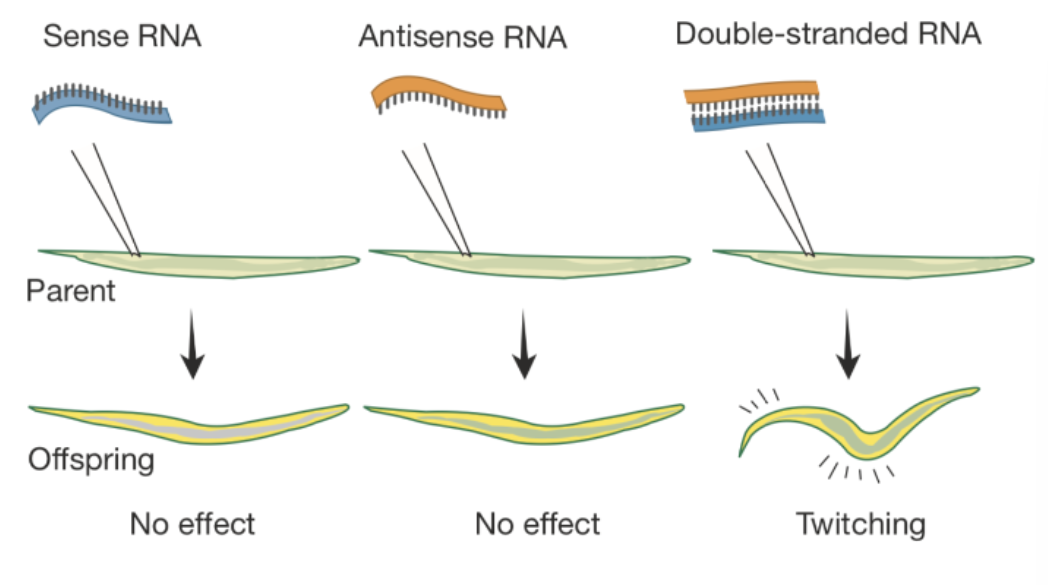
Describe Andrew Fire’s and Craig Mello’s 2006 experiment discovering RNAi in C. elegans.
RNA for a muscle protein is injected into C. elegans
When single-stranded RNA is injected, it has no effect
When double-stranded RNA is injected, the worm starts twitching in a similar way to worms carrying a defective gene for the muscle protein
Craig and Mello performed in situ hybridizations to confirm their results
Uninjected embryo → normal staining
Single-stranded antisense RNA → less staining but staining was still present
Double-stranded antisense and sense RNA → complete lack of staining
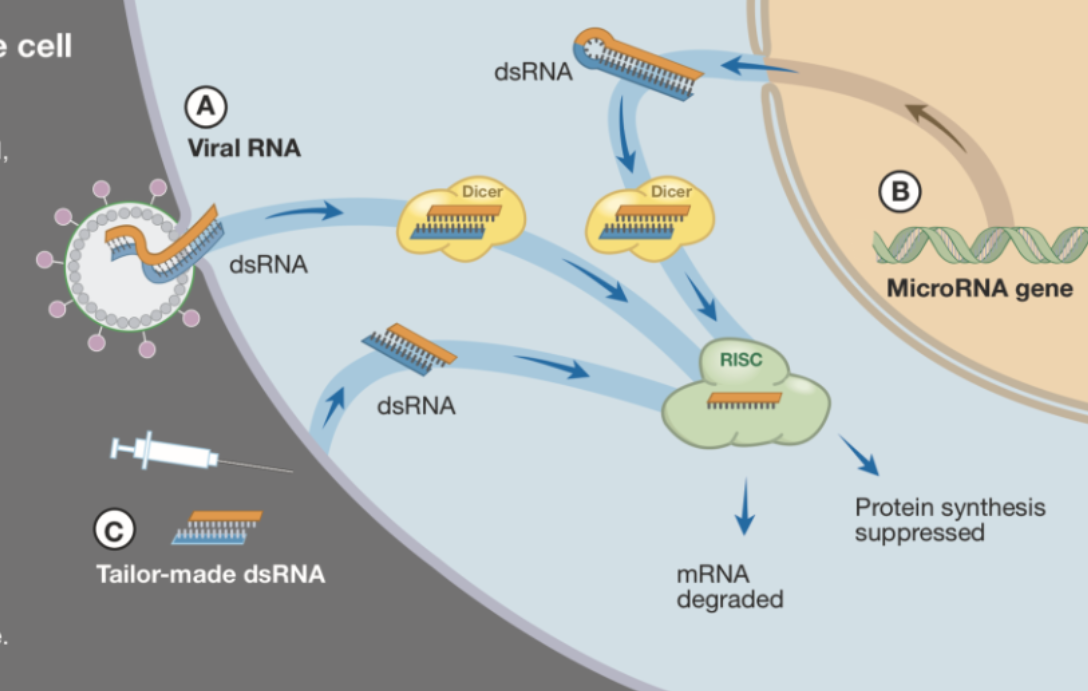
RNA interference (RNAi)
A biological process where siRNA and miRNA silence gene expression by targeting and degrading specific messenger RNA (mRNA)
General mechanism for RNAi:
Double-stranded RNA binds to the protein Dicer, which cleaves dsRNA into smaller fragments
One of the RNA strands binds to the RISC complex and hybridizes to the mRNA strand by base pairing
The mRNA is cleaved and destroyed, and no protein can be synthesized
Genetic screens use RNAi, introducing small non-coding RNA molecules through transfection or microinjection, in order to study mutant phenotypes!
Cell proliferation
Cell size
Live/dead
Gene expression
Cell behavior/morphology
Animal, tissue, cellular differentiation and development
Polarity and body axis
Wound healing

Compare siRNAs vs. miRNAs.
Origin and structure:
siRNA originates from mRNA or viral RNA and starts out as an RNA duplex or single-stranded RNA that forms long hairpins
siRNA starts out as a long double-stranded sequence that gets cleaved by Dicer into lots of small pieces
miRNA originates from mRNA and starts out as a single-stranded RNA that forms short hairpins of double-stranded RNA
miRNA starts out already small and forms a hairpin structure
Size:
siRNA and miRNA are 21-25 nucleotides long
Function:
siRNA → degradation of mRNA
miRNA → degradation of mRNA and inhibition of translation
Target
siRNA → genes from which they were transcribed (self-inhibiting)
miRNA → genes other than those from which they were transcribed (broader range)
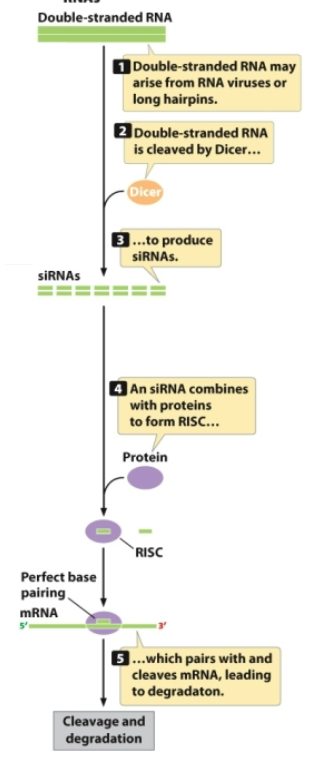
siRNA
A type of non-coding RNA that originates from mRNA or viral RNA, starts as an RNA duplex or ssRNA that forms long hairpins, degrades mRNA, and targets genes from which it was transcribed
siRNA originates from dsRNA that arises from RNA viruses or long hairpins
dsRNA is cleaved by Dicer to produce siRNAs
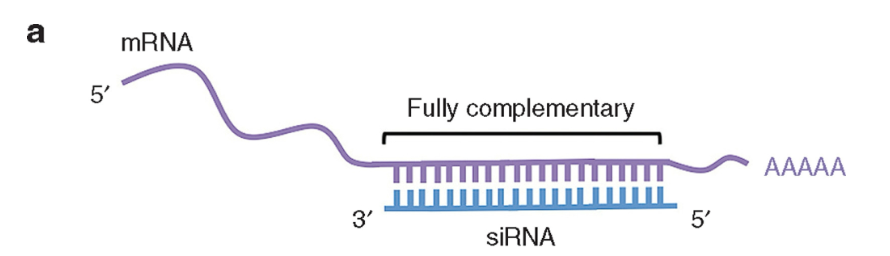
siRNA combines with proteins to form RISC complex → if there is a perfect base pairing with mRNA, the siRNA cleaves and degrades the mRNA
miRNA
A type of non-coding RNA that originates from mRNA, starts as ssRNA that forms short hairpins of dsRNA, degrades mRNA and inhibits translation, and targets genes other than those from which it was transcribed
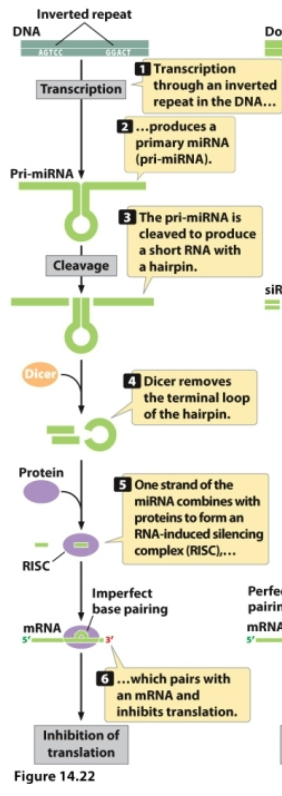
miRNA originates from transcription of DNA containing inverted repeats to produce a primary miRNA (pri-miRNA) shaped like a hairpin
pri-miRNA is cleaved to “cut” the hairpin out
Dicer cleaves off the terminal loop of the hairpin
One strand of the miRNA combines with proteins to form RISC complex → if there is an imperfect base pairing, the miRNA pairs with the mRNA and inhibits translation
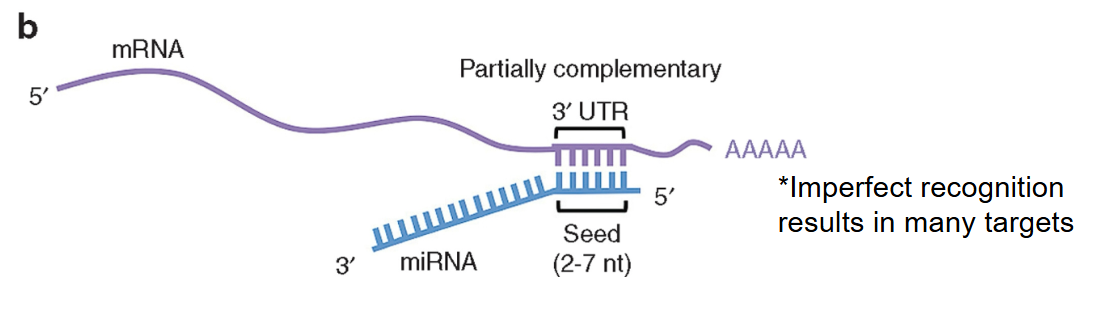
miRNA has a ___ range of targets than siRNA.
broader
Describe the significance and findings of Victor Ambros' and Gary Ruvkun’s 1993 discovery of microRNA.
The discovery of microRNA as a small non-coding RNA molecule capable of blocking mRNA translation was a seminal discovery, as it revealed a new dimension of gene regulation
Why are miRNAs important in gene expression and cellular differentiation?
A single miRNA can regulate the expression of many different genes, and a single gene can be regulated by multiple miRNAs, thereby coordinating and fine-tuning the entire network of a cell’s gene expression
miRNA regulation is universal among multicellular organisms!
Describe how RNA sequencing can be used to determine the expression of non-coding RNAs.
Isolate total RNA
Size-select for non-coding siRNA, miRNA, or piRNAs that are <30 nucleotides
Do not size-select for lncRNA
If you want lncRNA then do not apply a size filter, you will get both the lncRNA and mRNA sequences
Use random hexamers (primers) to generate cDNA
Not all siRNAs, miRNAs, or piRNAs have 3’ caps or poly(A) tails!
Oligo(dT) primers are specifically designed to be used for sequences with poly(A) tails, so you cannot use them here
Describe some examples of processes in the cell that use RNAi.
1) RNAi destroys viral RNA that has been injected into the cell, preventing the formation of new viruses.
2) miRNA regulates the synthesis of many proteins by preventing translation.
3) dsRNA molecules can be customized to activate the RISC complex to degrade mRNA for a specific gene.