Chapter 16: Regulation of Gene Expression
1/53
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
54 Terms
Gene expression
Gene expression is a very complex multi-step process and cells control which genes need to be expressed at what time, at what location (cell type or tissue type) and in what amounts.
cells does not express all genes all the time
Transcriptional Regulation
- processes that directly control gene expression, determines which genes are transcribed into mRNA.
housekeeping genes
are needed by almost every type of cell and appear to be unregulated or constitutive
Regulated genes
may or may not be expressed at any given time or in a given cell type
Gene Expression in Bacteria Can Be Regulated at Three Levels
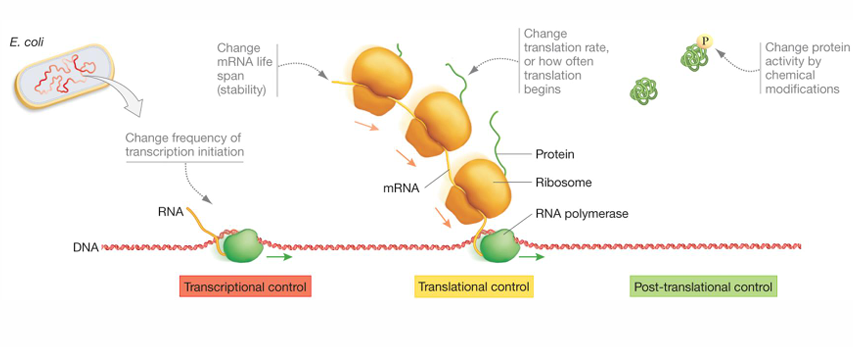
Transcriptional control
is particularly important due to its efficiency—it saves the most energy for the cell, because it controls gene expression before the cell expends many resources.
Translational Control
allows more rapid changes than transcriptional control in the amounts of different proteins because the mRNA has already been made and is available for translation.
Post-Translational control
provides the most rapid response of all three mechanisms because only one step is needed to activate or inactivate an existing protein
Which is the More Preferred mode of Regulation?
Transcriptional control is slow but efficient in resource use; posttranslational control is fast but energetically expensive.
Bacteria often respond to environmental change by
regulating transcription
Example: When lactose is present, E. coli makes enzymes to digest it – when lactose is absent, it does not make those enzymes.
operon
is a cluster of prokaryotic genes and associated regulatory sequences (DNA sequences involved in the regulation of a gene or genes). ▪Regulatory proteins bind to these regulatory sequences to control transcription of the genes. ▪One regulatory DNA sequence is the promoter – the site to which RNA polymerase binds to begin transcription.
Prokaryotic cells undergo
rapid alterations in biochemical pathways that allow them to adapt quickly to changes in their environment.
How are bacterial genes typically organized?
they are organized into physically linked structures called operons.
a transcriptional unit that includes transcription start/stop signals and usually regulatory elements and translation-specific sites.
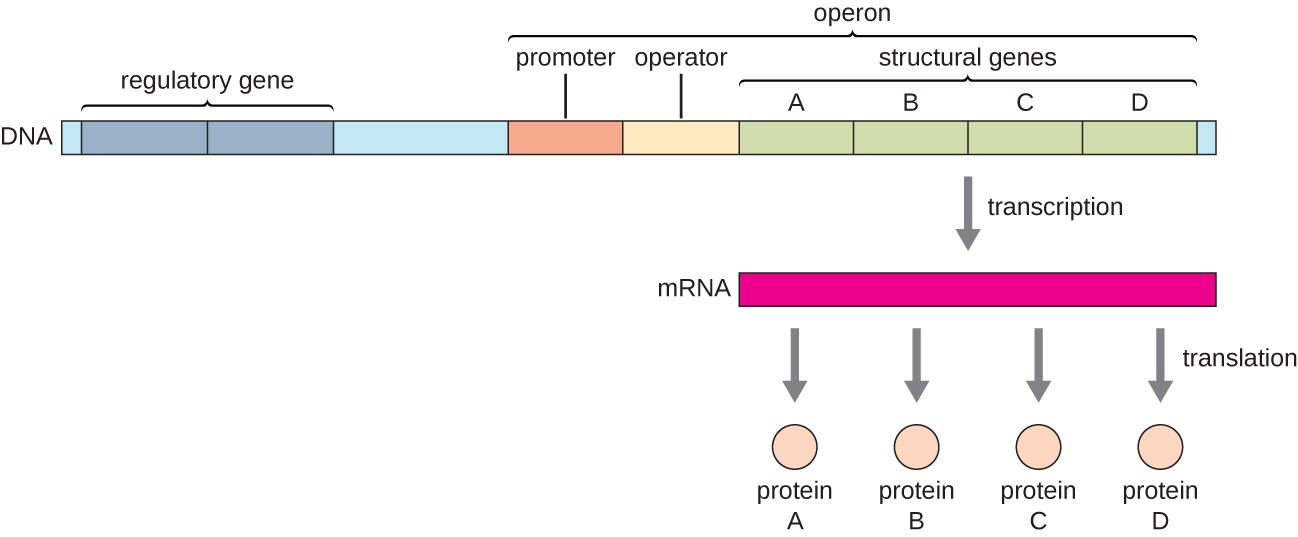
What is the difference between monocistronic and polycistronic operons?
Monocistronic operon = one gene; polycistronic operon = multiple related genes transcribed together.
What is a cistron?
A DNA segment that encodes a single polypeptide; in a polycistronic operon, multiple cistrons are regulated by one promoter.
What is an operator?
A repressor binding site where a trans-acting factor (repressor) binds to regulate RNA polymerase, often controlled by corepressors or inducers.
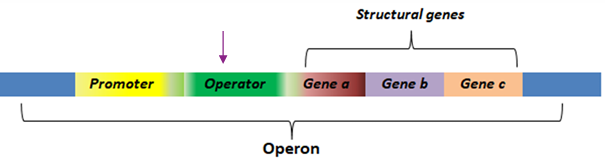
What is the role of the promoter in an operon?
The promoter region binds RNA polymerase to begin transcription.
lac operon
is a set of genes in E. coli (bacteria) that enables the cell to break down lactose when it is available.
No lactose → operon OFF
The lac repressor binds the operator and blocks RNA polymerase. No enzymes are made.Lactose present → operon ON
A lactose derivative (allolactose) binds the repressor, making it fall off.
RNA polymerase can now transcribe the operon → enzymes to digest lactose are produced.
What is an inducible operon?
An operon in which gene expression is turned on when a specific substance (e.g., lactose) is added to the environment.
What is a repressible operon?
An operon that is on by default but can be turned off by a small molecule (e.g., tryptophan).
How many structural genes does the lac operon contain, and what controls them?
Three structural genes, all under the control of a single promoter.
What does lacZ encode and what does it do?
Encodes β-galactosidase, which breaks lactose into glucose + galactose and converts lactose to allolactose.
What does lacY encode and what is its function?
Encodes lactose permease, a transmembrane pump that brings lactose into the cell and increases cell permeability to lactose.
What does lacA encode and what is known about its function?
Encodes β-galactoside transacetylase, which transfers an acetyl group from acetyl-CoA to thiogalactosides; its function is not well understood.
Which lac operon genes are essential for lactose catabolism?
lacZ and lacY only.
Positive Regulation of the lac Operon
Lactose converted to the inducer, allolactose, which inactivates Lac repressor.
Active adenylyl cyclase synthesizes cAMP to high levels. cAMP binds to activator CAP, activating it. Activated CAP binds to CAP site in the promoter.
RNA polymerase binds efficiently to the promoter.
Genes of the operon are transcribed at high levels.
Translation produces high amounts of proteins
Negative regulation of lac operon (lactose absent in the medium)
a. Lac repressor protein encoded by LacI gene binds to operator.
b. RNA polymerase is unable to bind to the promoter.
c. No transcription of the structural genes (Lac ZYA).
What happens in negative regulation of the lac operon (lactose absent)?
LacI repressor binds operator → RNA polymerase cannot bind → no transcription of lacZYA.
When lactose is present and glucose absent, what molecule inactivates the lac repressor?
Allolactose binds to repressor → changes shape → repressor falls off operator → transcription occurs.
How is lactose converted into allolactose?
By β-galactosidase (lacZ).
Allolactose binds to lac repressor at an allosteric site, changes its 3D conformation.
Lac repressor cannot bind to the operator.
RNA polymerase binds to the promoter and transcribes the structural genes (Lac ZYA).
What activates CAP?
cAMP (produced when glucose is absent).
Activated CAP binds promoter → increases transcription.
ATP is converted to cAMP by adenylate cyclase.
cAMP is a starvation signal. c. cAMP binds to CAP (catabolite activator protein) and activates it.
CAP binds to CAP binding site in the promoter stabilizing the RNA polymerase increasing the efficiency of transcription.
What happens when lactose and glucose are both present?
: cAMP levels low → CAP inactive → RNA polymerase binds weakly → very low transcription.
Differences between lac operon and trp operon?
Operon | Default State | Turns Off When | Type |
|---|---|---|---|
Lac Operon | OFF | No lactose | Inducible |
Trp Operon | ON | Tryptophan present | Repressible |
What type of operon is the trp operon?
Repressible; turned off when tryptophan is present.
Tryptophan
acts as a corepressor, a regulatory molecule that activates the repressor to turn off expression of the operon (a type of negative gene regulation)
How does tryptophan act in the trp operon?
As a co-repressor; binds trp repressor → activates it → repressor binds operator → stops transcription.
What happens when tryptophan is absent?
Repressor inactive → cannot bind operator → transcription of trpE–A occurs.
How is eukaryotic gene expression different from prokaryotic?
More complex; transcription in nucleus, translation in cytoplasm; multiple regulation levels.
Eukaryotic gene expression.
Eukaryotic chromosomes are found organized into chromatin.
Regulation is more complex than in prokaryotes.
Transcription and translation are spatially (by nuclear membrane) and temporally separated.
Regulated at transcriptional, post-transcriptional, translational, and post-translational levels.
Two types of chromatin.
i. Euchromatin – decondensed chromatin; lightly stained; active genes.
ii.Heterochromatin – Highly condensed chromatin; darkly stained; inactive genes.
i. Euchromatin
– decondensed chromatin; lightly stained; active genes.
ii.Heterochromatin
Highly condensed chromatin; darkly stained; inactive genes.
What is transcriptional regulation in eukaryotes?
Transcriptional regulation is the control of gene expression by regulating access of genes to RNA polymerase.
This is mainly achieved by altering chromatin structure so transcription machinery can (or cannot) reach the DNA.
It includes chromatin remodeling, histone modification, and DNA methylation.
What is chromatin remodeling and why is it important for transcription?
Chromatin remodeling is the process of opening or closing chromatin so genes can become accessible or inaccessible to transcription factors and RNA polymerase.
Two major processes contributute:
Physical movement of nucleosomes by ATP-dependent chromatin remodeling complexes.
Epigenomic (epigenetic) alterations, including chemical modifications of DNA and histone tails (histone code).
These modifications control whether chromatin is open (active) or closed (inactive).
What are the major epigenetic alterations to histones?
Epigenetic modifications include adding chemical groups to the amino-terminal histone tails:
Acetyl groups added to lysine
Methyl groups added to lysine or arginine
Phosphate groups added to serine or threonine
Ubiquitin added to lysine
These marks affect chromatin structure and gene accessibility.
Histone Acetyl Transferases (HATs):
Add acetyl groups (negatively charged) to lysine on histone tails.
Since DNA is also negatively charged, acetylation causes charge repulsion, loosening chromatin → gene ON.
Histone Deacetylases (HDACs):
Remove acetyl groups, causing chromatin to tighten → gene OFF.
What is DNA methylation and how does it affect gene expression?
DNA methylation involves adding methyl groups to cytosine residues in CpG islands using DNA methyltransferases.
Genes with highly methylated CpG islands are silenced (not transcribed).
Demethylases remove methyl groups to reactivate genes.
What is the organization of eukaryotic genes?
Exons (protein-coding)
Introns (non-coding)
Upstream regulatory regions include:Promoter, where transcription factors + RNA polymerase II bind to form the initiation complex
TATA box, which binds specific transcription factors
Promoter-proximal regions with tissue-specific activators or repressors
Enhancers, located far from the promoter, which bind activators and increase transcription
Enhancers loop to promoters using coactivators, increasing transcription initiation.
What are silencers and how do repressors work?
Silencers are DNA sequences where repressors bind to inhibit transcription.
Repressors can block transcription in three ways (not listed in your text, but typically by blocking activators, blocking RNA polymerase, or altering chromatin).
What is cell-specific gene expression?
Different cells express different genes depending on their needs.
Example: Human β-interferon is expressed only in virus-infected cells, demonstrating regulated, cell-specific transcription.
What is coordinated gene regulation?
Multiple genes can be activated simultaneously because they share the same regulatory sequence.
Example: Steroid hormones activate sets of genes that have the same hormone-response elements.
What is post-transcriptional gene regulation?
miRNAs (from endogenous genes)
siRNAs (from external sources like viruses)
These RNA molecules silence gene expression through RNA-induced silencing complexes (RISC).
Steps:
Pri-miRNA (~70 nt) processed by Drosha → pre-miRNA
Exported to cytoplasm by exportin 5
Dicer removes hairpin → double-stranded RNA
One strand degraded; other binds complementary mRNA → mRNA degraded or translation blocked
What is translational regulation?
Controls how efficiently mRNAs are used to make proteins. Occurs during the cell cycle and development.
Mechanisms include:
Adjusting poly(A) tail length (longer tail → more translation)
Phosphorylation of ribosomal proteins, altering translation
High temperature, which denatures proteins and prevents proper folding, reducing translation.
What is post-translational regulation?
Regulates proteins after they are made.
Includes:
Chemical modification (adding/removing groups)
Processing inactive precursors → active proteins
Example: Pepsinogen → pepsin
Protein degradation through the ubiquitin-proteasome system
Damaged proteins tagged with ubiquitin
Digested in ATP-driven proteasome
Peptides → amino acids → recycled or used for energy