DNA REPAIR
1/24
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
25 Terms
DNA Repair (learning objectives)
Explain how proofreading by DNA polymerase III takes care of the majority of mutations that arise during replication of DNA.
Describe the uvrABC system for removal of pyrimidine dimers.
Explain how DNA photolyase works.
State how does the cell recognizes the older strand of a dsDNA.
Explain how and why Uracil-DNA glycosidase removes uracil produced by deamination of cytosine.
Describe the SOS response and state its importance.
Explain the general role recombination has in repairing DNA.
Describe the different types of DNA repair active in the cell.
(Table 24-5, Types of DNA Repair Systems)
mismatch repair
base-excision repair (examples of damaged bases)
nucleotide-excision repair (thymine dimers, big adducts)
direct repair
Mismatch Repair (MMR): “Fixing incorrectly paired bases.”
Because mismatch repair fixes wrong base pairs created during replication (e.g., G–T, A–C).
The system recognizes the “mismatch” and “repairs” it.
Base-Excision Repair (BER): “Cutting out a damaged base and fixing the DNA.”
A damaged base (like uracil, oxidized C, alkylated G) is excised by a DNA glycosylase → then repaired.
DNA: “DNA” “glyco”: sugar “ase: “performing an action”, “An enzyme that acts on (or removes) the sugar-linked part of DNA.”
Nucleotide-Excision Repair (NER): “Cutting out an entire damaged nucleotide (or patch of nucleotides) to repair the DNA.”
NER removes bulky lesions (thymine dimers, big adducts) by cutting out a whole chunk of nucleotides, not just one base.
Direct Repair (Direct Reversal) Etymology
Direct → from Latin directus, “straight, immediate”
Repair → “restore or fix”
Literal Meaning
“Fixing the damage directly without removing the base or nucleotide.”
Why this name?
Because this is the only repair that:
does not cut out the base
does not excise nucleotides
just chemically reverses the damage in place
Examples:Photolyase directly reverses thymine dimers (non-human organisms)
MGMT directly removes a methyl group from O⁶-methylguanine
So the repair is direct
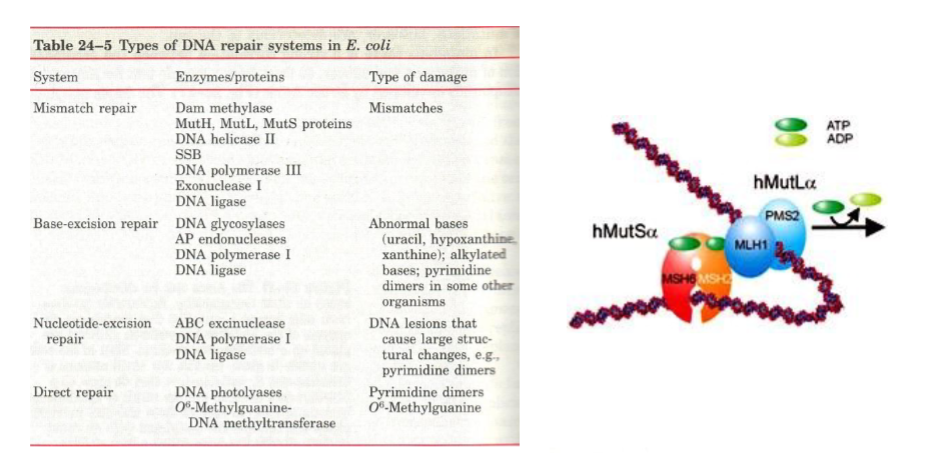
mismatch repair
Mismatch Repair (MMR): “Fixing incorrectly paired bases.”
I. MISMATCH REPAIR: For errors left by polymerase.
A. Methylation is a tag to identify the older strand. In E. coli, the Dam methylase marks the sequence GAMTC. This is needed to help decide which is the correct strand.
D = DNA
a = adenine
m = methyltransferase
This enzyme methylates adenine in DNA
1) Just after replication, the DNA is transiently hemimethylated by Dam Methylase. One methyl group is good for labeling up to 1 kb of DNA.
2) MutS binds to mismatched base pairs,
3) MutH binds to G AM T C. MutH acts as a site specific endonuclease, produces a nick 5' to the G.
Mut– = mutator (mutation-related gene/protein)
S = originally named “S gene product” from early screens
The letter does not stand for a meaningful word;
it is simply a designation from the original mutator strains.MutS is the mismatch recognition protein.
Mut– = mutator
H = again, a simple letter designation from early mutator experiments.
It does not stand for a specific biochemical term.MutH is an endonuclease activated by MutL and MutS, recognizes hemimethylated GATC sites
cuts the unmethylated strand (the newly synthesized one)
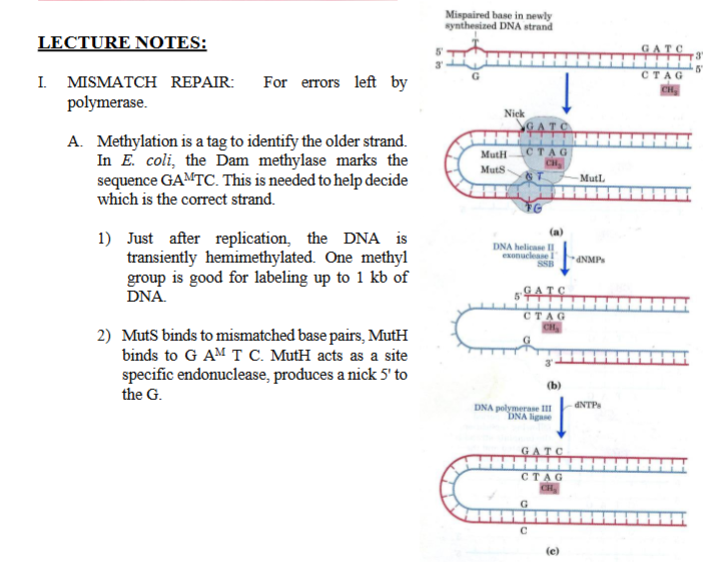
base-excision repair: the dna glycosylases
II. BASE-EXCISION REPAIR: THE DNA GLYCOSYLASES
DNA Glycosylases recognize the deamination of nitrogen bases (cytosine → uracil)
DNA glycosylase cleave the glycosyl bond (each base is attached to the sugar through a glycosyl bond, DNA glycosylase cuts this bond, removing the base only).
This leaves a hole (an AP site) which signals the cleavage site for AP nuclease. AP sites are normally formed slowly by spontaneous hydrolysis. However it is formed, the AP nuclease takes care of it ( AP endonuclease recognizes this “hole” and cuts the DNA backbone next to it, This creates an opening so DNA polymerase can insert the correct base)
AP = Apurinic / Apyrimidinic
= a site where a base is missing
B. Each glycosylase is specific for one type of lesion.
Ex: Uracil glycosylase removes uracil formed by deamination of cytosine. (deamination of cytosine → uracil)
C. Why does DNA has T instead of U? Because it would be impossible to distinguish a bonafide U from a deaminated C
.
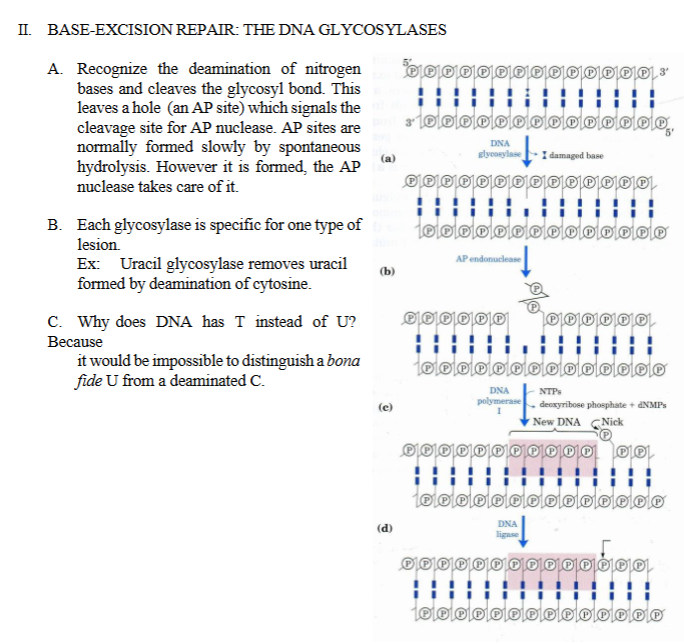
III. NUCLEOTIDE EXCISION REPAIR (bacteria repair)
III. NUCLEOTIDE EXCISION REPAIR - For distortions like thymine dimmer (damage from UV radiation)
NER removes a chunk of nucleotides instead of just one base.
ABC excinuclease is composed of 3 subunits encoded by uvrA, uvrB, and uvrC.
uvr = “UV repair.”
Excinuclease = a nuclease that removes (“excises”) a damaged segment of DNA.
ABC exinuclease makes 2 cuts around the distortion: 8 nucleotides 5' and 4 nucleotides 3'.
The ABC excinuclease doesn’t cut the damaged base itself.
Instead, ABC excinuclease cuts:
8 nucleotides upstream (5’ side)
4 nucleotides downstream (3’ side)
So ABC excinuclease removes a 12-nucleotide segment containing the damage.
In humans its 22-24 5' and about 5 at 3'.
Humans remove a larger patch during NER.
22–24 nucleotides on the 5’ side
~5 nucleotides on the 3’ side
Total removed in mammals = 27–30 nucleotides.
Much bigger than bacteria.
The 12 nucleotides strand is displaced by a helicase.
After excision:
A helicase unwinds and removes the damaged fragment.
This leaves a gap.
DNA polymerase I, and ligase, will finish the job.
Meaning:
DNA polymerase I synthesizes new DNA to fill the gap
Ligase seals the final nick in the sugar–phosphate backbone
This restores the DNA to normal.
In humans, DNA Pol δ/ε fills the gap.
Xeroderma pigmentosa patients lack this repair mechanism.
xero: “dry
derma; “skin”
pigemntosa: pigment “coloring matter”
Xeroderma Pigmentosum literally means: “Dry, pigmented skin.”
Xeroderma pigmentosum (XP) is a genetic disease where people cannot repair thymine dimers using NER.
Seven genes can cause X. pigmentosa.
There are seven possible XP genes (XPA through XPG).
Each one corresponds to a different protein in the NER pathway.
Mutations in any of these → XP disease.
Cell fusion and complementation assays can distinguish the defective gene.
If you fuse:
a cell from XP patient A
with a cell from XP patient
…and the fused cell repairs UV lesions, then:
Patient A and patient B had mutations in different XP genes.
If fusion does not restore repair:
Both patients have mutations in the same XP gene.
This is how scientists determine which XP gene is defective.
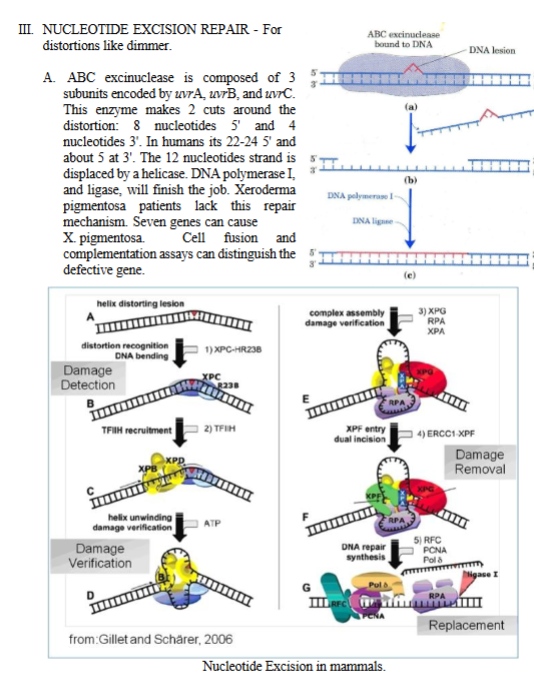
III. NUCLEOTIDE EXCISION REPAIR (mammals), look at the big picture on the bottom
1. XPC–HR23B: First responder that sees a distortion in the helix and marks the site for repair.
Meaning: Xeroderma Pigmentosum, Complementation group C, Human Homolog of Rad23, isoform B
TFIHH is recruited by XPC–HR23B
Transcription Factor II H
Function: Helicase + kinase complex that opens DNA around the lesion in NER, Contains XPB and XPD helicases.
2. TFIIH: A large transcription factor complex that contains two helicases:
XPB
XPD
TFIIH is brought to the lesion to begin unwinding the DNA around the damage.
3) Helicase activity
XPB (3’→5’ helicase) XPD (5’→3’ helicase)
Both use ATP to unwind ~20–30 nucleotides around the lesion.
DNA opens so the damaged base can be inspected.
XPD verifies the lesion is real and not a false signal.
4) Full assembly of repair complex (XPA,RPA, and XPG)
Proteins added:
XPA – verifies damage + helps position endonucleases (Xeroderma Pigmentosum group A protein)
RPA – binds to the undamaged strand to stabilize it (Replication Protein A)
XPG – Cuts DNA 3′ to the lesion (Xeroderma Pigmentosum group G protein)
XPA + RPA + XPG assemble around the lesion to prepare for dual incision.
5) XPG (Cuts on the 3′ side of the lesion) and ERCC1–XPF (Cuts on the 5′ side of the lesion) make two cuts, excising the damaged section, they remove a ~27–30 nucleotide single-stranded segment containing the damage.
6) RFC, PCNA, DNA Polymerase δ/ε
RFC (Replication Factor C) loads PCNA (the sliding clamp).
Pol δ or Pol ε fill in the missing nucleotides using the undamaged strand.
Polymerase synthesizes new DNA to replace the excised segment.
7) Ligase I: Seals the final nick, completing repair.

XPC–HR23B XPC etymology (nucleotide-excision repair in mammals)
1. XPC–HR23B XPC
XP = Xeroderma Pigmentosum (disease where NER fails)
C = Group C
Protein mutated in XP-C patients.
HR23B
H = human
R = RAD23 (yeast DNA repair protein)
23 = homolog of RAD23
B = isoform B (there is also HR23A)
Human protein similar to yeast RAD23; helps XPC stabilize and bind DNA.
TFIIH TF etymology (nucleotide excision base repair)
2. TFIIH TF = Transcription Factor II = RNA Polymerase II H = “H” complex (historical naming within TFII complexes)
“Transcription factor H for RNA polymerase II.”
A general transcription factor, later discovered to also function in NER.
XPB and XPD XP (etymology and nucleotide excision repair in mammals)
3. XPB and XPD XP = Xeroderma Pigmentosum B / D = Complementation groups B and D
Proteins named after XP patients whose cells were defective in NER.
XPB and XPD are helicases mutated in XP-B and XP-D patients.
XPA / XPG XP etymology (nucleotide excision repair in mammals)
4. XPA / XPG XP = Xeroderma Pigmentosum A / G = Complementation groups A and G
Same reasoning as above.
XPA = damage verification
XPG = 3′ endonuclease
The proteins that are defective in XP-A and XP-G patients.
RPA etymology (nucleotide excision repair in mammals)
5. RPA R = Replication P = Protein A = A subunit (largest), but the whole complex is called RPA
Also called Replication Protein A.
Protein involved in replication, identified originally by fraction A.
A single-stranded DNA–binding protein that stabilizes unwound DNA.
ERCC1–XPF ERCC1 etymology
6. ERCC1–XPF
E = Excision
R = Repair
C = Cross-Complementing
C1 = Complementation group 1
“Excision Repair Cross-Complementing group 1.”
Named because the human gene complemented a repair-defective hamster cell line.
XPF
XP = Xeroderma Pigmentosum
F = Complementation group F
ERCC1-XPF = the 5′ endonuclease that cuts DNA during NER
PCNA etymology (nucleotide excision repair mammals)
8. PCNA P = Proliferating C = Cell N = Nuclear A = Antigen
Originally discovered as a nuclear antigen in rapidly dividing cancer cells.
“Proliferating Cell Nuclear Antigen.”
Sliding clamp that surrounds DNA and increases polymerase processivity.
DNA polyermase I deta and episolon (nucleotide excision repair)
9. DNA Polymerase δ (delta) & ε (epsilon) Polymerase
From Greek poly- (many) + meros (parts)
→ “enzyme that makes many-parts (polymer)."
Greek Letters
δ (delta) = fourth letter
ε (epsilon) = fifth letter
These names reflect the order of discovery within eukaryotic polymerases.
DNA Pol δ/ε synthesize new DNA in the NER gap.
DNA Ligase I etymology (nucleotide excision repair mammals)
10. DNA Ligase I Ligase
From Latin ligare = “to tie, bind, fasten.”
Enzyme that ties (ligates) the DNA backbone back together.
DIRECT REPAIR
what does direct repair do?
DNA photolyases (300-500nm, FADH2 and folate)
O⁶-Methylguanine DNA Methyltransferase (MGMT)
IV. DIRECT REPAIR - Repairs several types of damage. Does not remove bases or nucleotides
Direct repair =
DNA damage is fixed directly, by reversing the chemical modification.
No:
Base removal
Nucleotide removal
Cutting of DNA
This is different from BER or NER.
A. DNA photolyases. For repair of cyclobutane pyrimidine dimers, produced by UV light
Found in all cell types and in all life forms. Binds to T:T dimers and absorbs light 300-500 nm Then, it breaks the dimmer. Two cofactors (or chromophores) are needed: FADH2 and folate.
DNA: DNA
Photo: light
lyases: “to split apart”
Photolyase uses:
FADH₂ = reduced flavin adenine dinucleotide
MTHF (folate) = methenyltetrahydrofolate
These are light-absorbing molecules (chromophores) that transfer the light energy to the enzyme’s active site.
They make photoreactivation possible.
B. 06 - Methylguanine DNA methyltransferase. Transfers a -CH3 group FROM GUANINE (undoing the damage) to a specific Cys amino acid. Not really an enzyme because the whole protein is used only once.
What does this enzyme fix? Alkylating agents sometimes place a methyl group (–CH₃) onto the oxygen at the O⁶ position of guanine.
This damaged base:
mispairs with thymine
→ causes G→A transitions
MGMT reverses this damage.

what are two ways thymine dimers are repaired?
ner and direct repair (dna photolyase)
V. TRANSCRIPTION COUPLED REPAIR (TCR)
V. TRANSCRIPTION COUPLED REPAIR (TCR)
The transcribed DNA strand is repaired faster than the non transcribed strand.
TCR is activated when the RNA polymerase stops at a deformity (damage). RNA polymerase was transcribing.
The stopping of RNA polymerase recruits the excision repair enzymes mentioned above.
The gene for susceptibility to breast and ovarian cancer, BRCA1, encodes a zinc finger protein, that is phosphorylated and relocated into the nucleus in response to DNA damage.
BRCA1 interacts with RAD51 to carry out TCR. In sum, altered TCR is responsible for accumulation of mutations causing many breast and ovarian cancers (see Science, below
VI. Heavy Damage (SOS response (emergency system)
SOS response (emergency system)
ssDNA
RecA
LexA
umuC, umuD (DNA Pol V)
VI. HEAVY DAMAGE
Is dealt with by error-prone repair mechanisms. This is an emergency system and the aim is just to stay alive (better mutated than dead). This is called the SOS response.
When DNA is severely damaged (UV, chemicals, double-strand lesions), normal accurate repair systems cannot keep up.
If the cell waits for perfect repair, it will die.
So the cell activates an emergency survival system, called the: SOS Response = “Better mutated than dead.”
Here is the full SOS logic:
DNA damage → ssDNA accumulates
RecA binds ssDNA → becomes RecA*
RecA* induces self-cleavage of LexA
LexA repression lifts → SOS genes turn ON
Early SOS genes: UvrABC (accurate NER)
Late SOS genes: umuC/umuD → Pol V (error-prone, mutagenic)
Cell repairs DNA enough to survive (even with mutations)
SOS response → bacterial emergency DNA repair system
ssDNA → distress signal
RecA → activates SOS by cleaving LexA
LexA → repressor that keeps SOS genes OFF
umuC/umuD → Pol V → error-prone polymerase used as a last resort

VII. Suicide or Apoptosis (human cells)
eukaryotes
p53
how does p53 monitor the extend of DNA damage in humans?
what does p53 do if there is too much of a mismatch?
VII. Suicide or Apoptosis
When DNA damage is very severe, human cells do not use error-prone DNA synthesis like bacteria.
Instead: The cell chooses to commit suicide (apoptosis) to protect the entire organism.
This prevents damaged cells from:
becoming cancerous
passing on mutations
disrupting tissues
This response is controlled by p53, a tumor suppressor protein
Suicide or Apoptosis is the response of the higher eukaryotes cells to heavy damage.
p53 monitors the extent of DNA damage in humans by inspecting recombination making sure that the newly formed heteroduplex DNA matches. p53 inhibits recombination if there is too much mismatch. In this case, p53 induces the program for cell death, or apoptosis.
Ultra-Simple Summary
p53 monitors DNA damage.
It checks whether recombination repair is accurate.
If mismatches are too high → recombination is unsafe.
p53 decides the cell is too damaged to fix.
p53 activates apoptosis → controlled cell suicide.
This prevents mutations from turning into cancer

VIII. Repair of double strand breaks (DSB)
two major pathways: 1) general recombination 2) illegitimate recombination (or end joining)
Werner’s Syndrome
VIII. Repair of double strand breaks (DSB)
A double-strand break (DSB) is one of the most dangerous forms of DNA damage because both strands of the helix are broken.
If not repaired, the chromosome can be lost or rearranged.
The cell has two major pathways to fix DSBs:
1) General recombination and 2) Illegitimate recombination (or end joining).
1) General Recombination (Homologous Recombination, HR)
Uses a sister chromatid as a template
Accurate (high fidelity)
Requires proteins like RAD51, BRCA1, etc.
Occurs mostly during S and G2 phases (when sister chromatids are present)
This is the “good,” accurate repair pathway.
2) Illegitimate Recombination (End Joining, NHEJ)
Also called Non-Homologous End Joining
Joins the broken ends without a proper template
Very error-prone
Can add or delete nucleotides
Can create chromosomal translocations
Illegitimate Recombination is the emergency pathway when no accurate template is available
The steps of general recombination are depicted in fig. 2, and those of the illegitimate recombination are shown in fig. 3.
DNA Repair processes, and specially the repair of DSB, are error prone. This means that an individual with an increased rate of repair will accumulate mutations and thus will age faster. These individuals have a greater chance of developing diseases typical of old age. For example, most cases of Werner’s Syndrome lack a topoisomerase needed to unwind the DNA. Replication will cause DSB because the structural tension of the unwinding DNA can not be alleviated. Two cases of Werner’s Syndrome are shown. Typically the symptoms become evident in adolescence.
“The syndrome first described by Werner, consisting of a characteristic set of symptoms.”
Werner syndrome is a premature aging disorder

VIII. Repair of double strand breaks (DSB)
two major pathways: 1) general recombination 2) illegitimate recombination (or end joining)
1⃣ MRX/MRN complex proteins
2⃣ Homologous recombination proteins (Rad family)
3⃣ Mismatch repair proteins (MutS / MutL homologs)
1. MRX / MRN COMPLEX (DSB repair)🔹 RAD50Etymology
RAD = Radiation sensitive
50 = mutant #50 discovered in yeast screens sensitive to ionizing radiation
Long coiled-coil protein with zinc-hook domain
Provides ATPase activity
Acts as a tether to hold broken DNA ends together
🔹 MRE11 Etymology
MRE = Meiotic Recombination
11 = 11th mutant identified
Meaning / Function
Nuclease that performs 5’→3’ endonucleolytic trimming
Generates 3’ overhangs needed for Rad51 binding
Partner of Rad50 in the MRX complex
🔹 XRS2 (yeast)Etymology
XRS = X-ray sensitive mutant
2 = 2nd gene discovered in that complementation group
Meaning / Function
Scaffold protein that stabilizes Mre11/Rad50
Required for nuclear localization of the complex
Human homolog = NBS1 (Nijmegen breakage syndrome protein)
🧬 MRX complex (Mre11–Rad50–Xrs2)
Mre11 = nuclease
Rad50 = ATPase + DNA tether
Xrs2 = regulatory scaffold (human NBS1)
In humans, the same complex is MRN:
Mre11–Rad50–Nbs1
🧩 2. MRX + Sae2Sae2
Etymology:
SAE = Suppressor of Alpha-Endoprotease (historical yeast genetic screens)
Meaning / Function
Initiates end resection with Mre11
Helps remove protein blocks at DNA ends
Human homolog: CtIP
🧩 3. RAD51, RAD52, RAD54, RAD55, RAD57
(All discovered as RADiation-sensitive repair genes.)
🔹 RAD51Meaning / Function
Yeast homolog of RecA
Catalyzes strand invasion during homologous recombination
Central recombinase of the HR pathway
🔹 RAD52Meaning / Function
mediates Rad51 loading onto ssDNA
promotes annealing of complementary strands
🔹 RAD54Etymology: Radiation-sensitive gene 54 Function
Chromatin-remodeling ATPase
Enhances Rad51 strand invasion
Helps displace Rad51 after pairing is complete
🔹 RAD55 and RAD57Meaning / Function
Rad51 paralogs
Form Rad55–Rad57 heterodimer
Stabilize Rad51 filaments on ssDNA
Help overcome RPA competition
🔹 RPAMeaning
Replication Protein A
Function
Binds ssDNA
Stabilizes the DNA but initially blocks Rad51
Must be displaced by Rad52/Rad55–57 for strand invasion
4. Mismatch Repair (MutS & MutL homologs)🔹 Msh2Etymology
Msh = MutS homolog
2 = homolog #2 identified in eukaryotes
Meaning / Function
Core subunit of mismatch recognition
Pairs with Msh6 or Msh3 depending on mismatch type
🔹 Msh3Meaning
MutS homolog #3
Function
Forms MutSβ (Msh2–Msh3)
Recognizes large insertion–deletion loops, hairpins, microsatellite expansions
🔹 Msh6Meaning
MutS homolog #6
Function
Forms MutSα (Msh2–Msh6)
Recognizes single-base mismatches and small 1–2 bp loops
🧬 MutS complexes:
MutSα = Msh2 + Msh6 → point mismatches
MutSβ = Msh2 + Msh3 → insertion/deletion loops
🧩 5. MutL homologs: Mlh1 and Pms1🔹 Mlh1Etymology
Mlh = MutL homolog
1 = the first discovered human homolog
Meaning / Function
Scaffold protein → recruits endonucleases for mismatch excision
Major Lynch syndrome gene
🔹 Pms1Etymology
Pms = Post-meiotic segregation
(named based on mutant phenotype in yeast genetics)
Meaning / Function
Works with Mlh1 (Mlh1–Pms1 heterodimer = MutLα)
Coordinates strand discrimination and nicking of the error-containing strand
🧬 Putting it all together (super compact) Double-Strand Break Repair (HR):
Rad50 – structural ATPase, tethers ends
Mre11 – nuclease
Xrs2/Nbs1 – regulatory scaffold
Sae2/CtIP – initiates resection
Rad51 – recombinase
Rad52 – loads Rad51
Rad54 – chromatin remodeling
Rad55/57 – stabilize Rad51 filament
RPA – binds ssDNA until Rad51 displaces it
Mismatch Repair (MMR):
MutSα (Msh2–Msh6) – recognizes single-base mismatches
MutSβ (Msh2–Msh3) – recognizes larger loops
MutLα (Mlh1–Pms1) – coordinates cleavage and repair

VIII. Repair of double strand breaks (DSB)
two major pathways: 1) general recombination 2) illegitimate recombination (or end joining)
DOUBLE-STRAND BREAK (DSB) REPAIR — ILLEGITIMATE RECOMBINATION (NHEJ)
Illegitimate recombination = the NHEJ pathway, because it joins ends with no need for homology (“illegitimate” = no proper matching).
Step | What Happens | Key Proteins |
|---|---|---|
1. DSB occurs | “Lightning bolt” breaks DNA | — |
2. End recognition | Broken ends held together | Ku70/Ku80 |
3. Synapsis & activation | Recruits & activates repair machinery | DNA-PKcs |
4. End processing | Trimming, filling, repairing ends | Artemis, pol μ, pol λ |
5. Scaffold assembly | Align ends & stabilize ligase | XRCC4 |
6. Ligation | DNA ends sealed | Ligase IV |
7. Completion | DNA restored (mutagenic) | — |
1. Ku70 / Ku80 Etymology
Ku = named after the Ku antigen, discovered in a Japanese patient with autoimmune disease who produced antibodies against this protein.
70 & 80 = molecular weights (70 kDa and 80 kDa).
Meaning / Function
Heterodimer that clamps around broken DNA ends.
Protects the ends + recruits the entire NHEJ machinery.
“Ku” = autoimmune antigen; 70/80 = sizes.
2. DNA-PKcs Full name: DNA-dependent Protein Kinase catalytic subunit Etymology
DNA-dependent: requires DNA to activate
Protein kinase: phosphorylates proteins
cs = catalytic subunit (large active part of the enzyme)
Meaning / Function
Activated by Ku70/Ku80.
Creates the DNA-PK holoenzyme.
Phosphorylates Artemis + other NHEJ proteins.
“DNA-PKcs” = the kinase that turns ON when bound to DNA.
3. Artemis Etymology
Named after Artemis, the Greek goddess of the hunt (symbolizing “cutting”).
Mutation causes RS-SCID (Radiation-sensitive severe combined immunodeficiency).
Meaning / Function
Endonuclease activated by DNA-PKcs.
Trims incompatible DNA ends.
“Artemis” = nuclease that cuts DNA ends.
4. Pol μ (DNA Polymerase mu) & Pol λ (DNA Polymerase lambda) Etymology
Greek letters:
μ (mu)
λ (lambda)
Names given based on polymerase family classification.
Meaning / Function
Pol μ → fills gaps with minimal template; very error-prone.
Pol λ → more accurate but still used in NHEJ.
These are “translesion-style” polymerases made for broken ends.
Greek letters simply denote polymerase family subtypes.
5. XRCC4 Full name: X-Ray Cross-Complementing 4 Etymology
XR = X-Ray
CC = Cross-Complementing (functional grouping of mutant strains)
4 = the 4th gene in the complementation group
Named because cells lacking XRCC4 were hypersensitive to X-ray–induced DNA damage.
Meaning / Function
Scaffold protein that binds and stabilizes Ligase IV.
Helps align DNA ends for ligation.
“XRCC4” = gene discovered through X-ray sensitivity screens.
6. DNA Ligase IV Etymology
Ligase = enzyme that ligates (joins) DNA ends
IV = the 4th ligase type identified in mammals
Meaning / Function
Seals the final phosphodiester bond in NHEJ.
Works even when ends do NOT match → causes mutations.
“Ligase IV” = the NHEJ ligase.
BONUS: Other NHEJ-associated factors (brief meanings) XLF (Cernunnos)
Cernunnos = Celtic god whose antlers “link things together.”
Meaning: XRCC4-like factor; helps ligation.
PAXX
Protein Associated with Xrcc4-like X factor.
CtIP (human homolog of Sae2)
C-terminal binding protein–interacting protein
Early end-resection regulation.

Werner Syndrome
Werner syndrome is a premature aging disorder (“progeria of adults”).
topoisomerase disorder → too much tension → DNA snaps → double stranded breaks → cells gets too many DSBs → premature aging
