Gene Control
1/46
There's no tags or description
Looks like no tags are added yet.
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
47 Terms
Cell commitment
Even without phenotypic expression cells are committed to a particular differentiated state
DNA is ____ but _____ expressed
Unchanged, Differentially
Purpose of a Northern blot
Detects specific mRNA and is used to check the expression of a gene
uses a probe
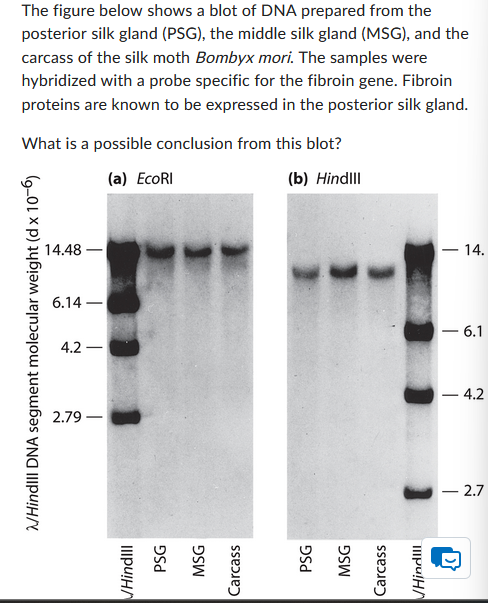
Southern Blot
Probe detects specific DNA.
Tandem Mass Spectrometry (How does MS/MS)
Used for peptide sequencing. Peptide is fragmented into smaller ions, and the mass differences between consecutive fragment ions reveal the amino acid sequence
RT-PCR
Synthesizes cDNA or complementary DNA from an RNA template that is amplified using PCR
Nuclear Run On
measure active gene transcription by allowing RNA polymerases in isolated nuclei to extend nascent RNA strands with labeled nucleotides. The labeled RNA is then hybridized to gene-specific probes to identify which genes were being transcribed at the time of isolation.
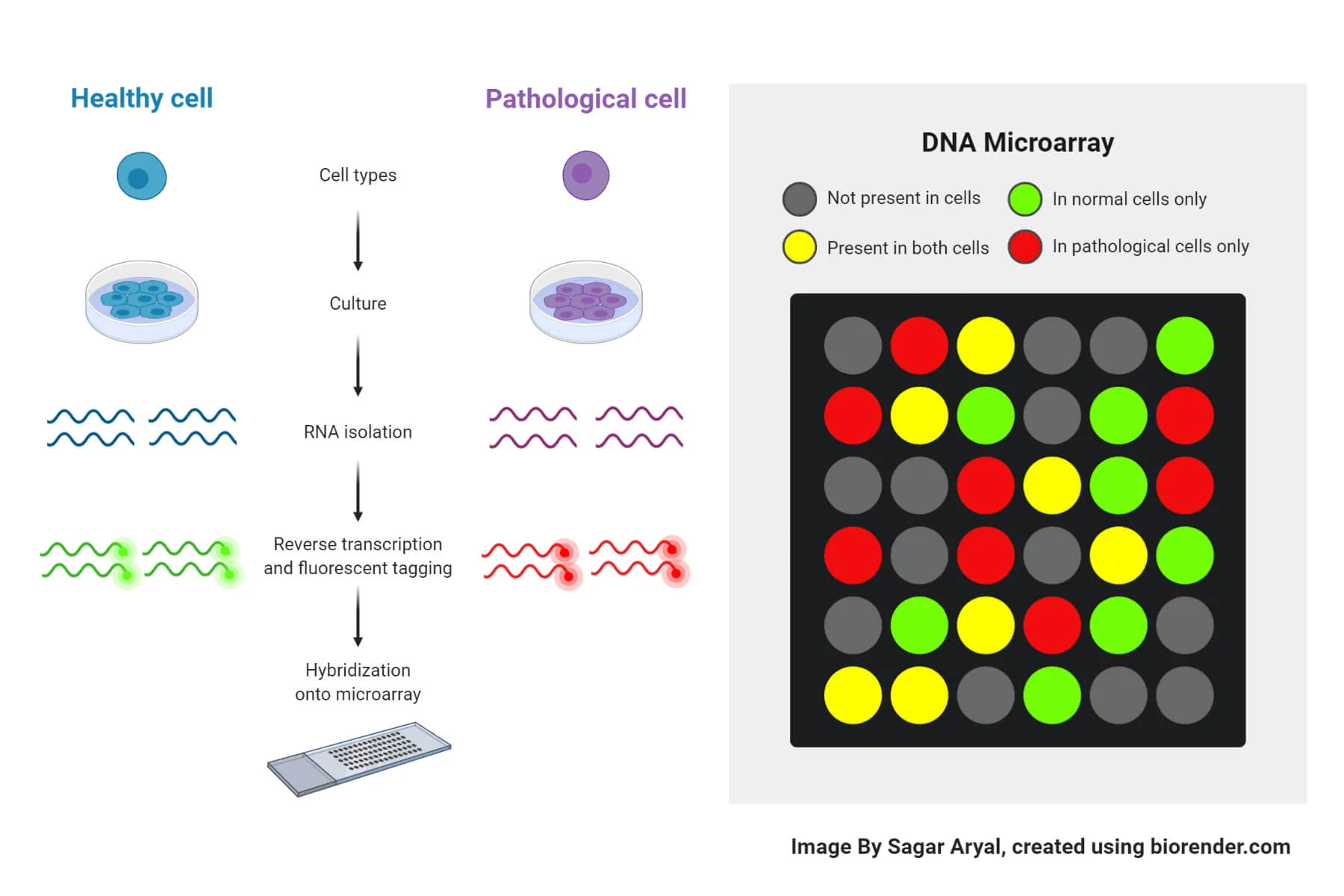
DNA microarray
used to compare gene expression under different conditions
ex. comparisons of gene expressions in diseases vs normal cell
miRNA
Regulates gene expression post-transcriptionally by binding to complementary sequences on mRNAs, leading to degradation or translational repression.
single stranded RNA
20-25 base pairs
siRNA
double stranded
Defends against foreign nucleic acids (like viruses, transposons) by guiding the cleavage and degradation of target mRNAs.
Silences transposable elements in germ cells and helps maintain genome stability.
21-31 double strands
lncRNA
200 bp long regulatory RNA
bidirectional promoter
Regulates gene expression at multiple levels:
chromatin modification → repression
transcription:
competes for transcription regulatory proteins → repression
recruit transcription activator → activator
post-transcriptional inhibition
Xist lncRNA → coats one X chromosome in females to inactivate it (X-chromosome inactivation).
Steps of RNA Transcription
Initiation
Elongation
Termination
Promoter
Where RNA polymerase first binds
Important Sequences of the Promoter
-35 and -10
aka. consensus sequence
-35 = TTGACA
-10 = TATAAT
What is needed for RNA Polymerase to recognize the -35 and -10 sequences?
sigma factor
Transcriptional start site
+1
Elongation (transcription)
The chaining of RNA Nucleotides together
rNTP enter and start chaining together
nucleotide is added to the 3’ end of the RNA
Kinetic Proofreading
Involves pyrophosphate lysing the bond or something of the RNA
Nucleolytic proofreading
involves nuclease within RNA polymerase to lyse the bond of the RNA
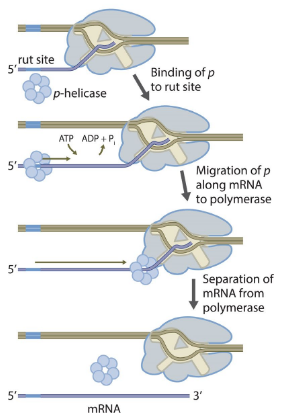
Rho Dependent Termination (transcription)
A termination mechanism that requires the Rho protein, a helicase.
Rho binds to a rut site (Rho utilization site) on the nascent RNA.
It moves along the RNA (using ATP) toward the RNA polymerase.
When RNA polymerase pauses at a termination sequence, Rho catches up.
Rho unwinds the RNA-DNA hybrid inside the transcription bubble, releasing the RNA transcript.
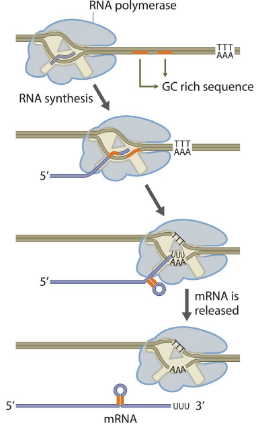
Rho-Independent Termination (Transcription)
The RNA transcript forms a GC-rich hairpin loop followed by a series of uracils (poly-U tail).
The hairpin causes RNA polymerase to pause.
The weak A–U base pairing between RNA and DNA makes the RNA-DNA hybrid unstable.
The RNA transcript dissociates, ending transcription.
tRNA
the link between the mRNA and the polypeptide
is in a three leafed clover shape
tRNA synthetase
add amino acids to the tRNA by an active site.
charges the tRNA
amino acids are attached by their carboxyl group to the 3’OH of the tRNA
Structure of Ribosomes
2/3 RNA, 1/3 protein
Contains a large ribosomal subunit and a small ribosomal subunit
Ribozyme
all enzymatic activity in the ribosome is by the RNA components = ribozyme
Binding sites of Ribosomes
A site
P site
E site
A site of Ribosome
aminoacyl site, binds an aminoacyl-tRNA
P site of ribosome
peptidyl site, binds a tRNA with an attached growing polypeptide chain
E site of ribosome
exit site, binds to deacylated tRNA that is to be released from ribosome
Translation Initiation
Small ribosomal subunit with translation initiation factor bind and then bind to mRNA
small ribosomal subunit with bound initiation tRNA moves along mRNA and searches for the first AUG
large ribosomal subunit binds
Charged tRNA binds to A site
firs peptide bond forms
need for gene regulation in bacteria
Control gene expression in response to environmental changes.
Energy conservation: Proteins made only when needed.
Types of genes:
Constitutive → always on (basic survival).
Regulated → active only under certain conditions.
Regulation types:
Temporal (right time)
Spatial (right place)
Quantitative (right amount)
Cis elements
DNA sequences located on the same molecule of DNA as the gene(s) they regulate.
Examples: Promoter, Operator, Enhancer/Silencer sequences.
Trans elements
Regulatory molecules (usually proteins or RNAs) that diffuse through the cell and act on cis elements.
Can regulate genes on other DNA
Examples: Repressor protein, Activator protein, Sigma factors.
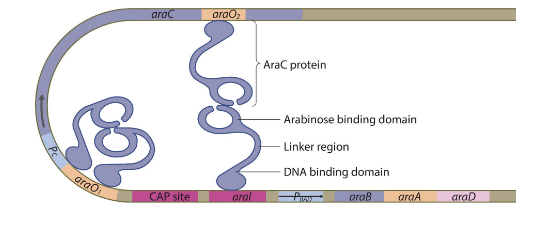
Ara operon in the absence of arabinose
the araC protein is made as usual but binds to each other to form a dimer and blocks the RNA polymerase from transcribing the genes
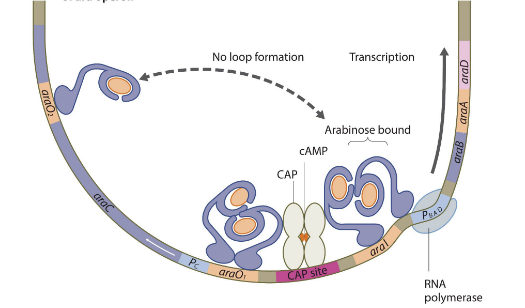
ara operon in the presence of arabinose
arabinose binds to the araC proteins so they loosen their grips on each other and move to adjacent areas near the promoter. Then CAP protein binds to the CAP site in the absence of glucose and activates transcription.
CAP protein in lac operon
enhances transcription. Needs cAMP to be activated and bind to the cAMP site.
In the presence of glucose, the CAP protein is INACTIVATED
best conditions in a lac operon is for lactose to be present but no glucose.
gene regulation by mRNA
Gene regulation at the mRNA level happens after transcription but before (or during) translation. It controls how much protein is made from the mRNA.
Riboswitch
RNA molecule acting as a switch depending on the effector molecule binding
main regulation in eukaryotes
🔴 Negative Regulation (Repressor-based)
Similar to bacterial negative inducible systems (e.g., lac operon).
Repressor proteins bind promoters → block transcription.
A signal (e.g., heat shock, stress) inactivates the repressor → gene is expressed.
🟢 Positive Regulation (Activator-based)
Many eukaryotic genes are default OFF.
Require activator proteins to initiate transcription.
Activators may:
Be tissue-specific.
Require cofactors (e.g., steroid hormones).
Common in higher organisms → ensures genes expressed only in correct tissue, time, or signal.
Summary:
Negative regulation = block lifted → gene ON.
Positive regulation = activator required → gene ON.
Front (Q): What is chromatin remodeling?
The process of altering nucleosome structure/position to regulate DNA accessibility.
Involves ATP-dependent remodeling complexes + histone modifications.
Switches chromatin between euchromatin (open/active) and heterochromatin (closed/silent).
What are the main mechanisms of chromatin remodeling?
Nucleosome repositioning/sliding → DNA exposed for transcription factors.
Histone modifications:
Acetylation → loosens chromatin → transcription ON.
Deacetylation → tightens chromatin → transcription OFF.
Methylation → context-dependent (ON or OFF).
Phosphorylation, ubiquitination: additional regulation layers.
Phosphorylation, acetylation:
→ add negative charges
→ neutralize basic histone
→ repel DNA or other modified histone
→ interaction with regulatory protein
micrococcal nuclease
Micrococcal nuclease digests exposed DNA between nucleosomes. Then, only purify the ones that have histones attached to it, getting rid of the histone protein parts and the DNA left will be sequenced.
Break in DNA means a histone was present in tumor vs normal cell
So, if you map all the cut sites across the genome, you can figure out nucleosome positioning:
Break in DNA sequence after MNase digestion = accessible, unprotected DNA (between nucleosomes).
No break (protected region) = nucleosome present.
3C, 5C experiment
Involves immunoprecipitation using chromatin
LCR
controls expression of all the globin genes
functions to keep open chromatin conformation
deletion leads to low globin gene expression - Hispanic thalassemia