Genomic studies, cloning and trangenic tech
1/25
Earn XP
Description and Tags
Name | Mastery | Learn | Test | Matching | Spaced |
|---|
No study sessions yet.
26 Terms
how to identify genes/mutations that are involved in diseases?
look for association of phenotype/ disease w known sequences in genome to identify disease associated regions/genes
gene related AMD
less than 50% of AMD cases can be attributed to mutations in genes
linkage
traits (SNPs, gnees, phenotypes) are linked if they are usually inherited together
2 traits that ARE physically CLOSE on a chromosomes have high chance of being inherited together (linked)
2 traits on diff chromosomes are NOT linked and have 50/50 chance of being inherited together
single nucleotide polymorphism (SNP)
single nucleotides within the genome that VARY between individuals
Often found outside of genes
used in mapping gene/disease location
why are SNP helpful in mapping disease/gene location?
they may locate near a potential candidate gene that is involved in disease/trait/phenotype under study
recombination
exchange of HOMOLOGOUS segments of chromosomes - mixes up alleles on sister chromosomes (crossover)
occurs AFTER DNA rep (when chromosomes are condensed and aligned)
occurs during meiosis (driving force for evolution in euk)
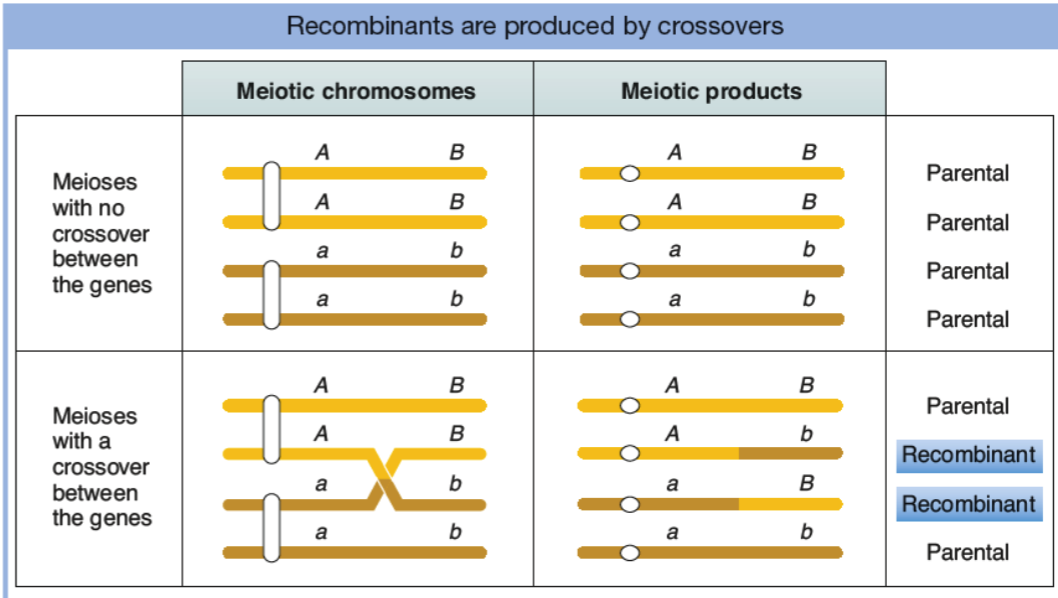
how is family linkage disease be identified?
approach:
identify proband (1st person who is diagnosed)
draw pedigree
isolate DNA from affected and unaffected members
identify specific SNP alleles present in each subject across the genome (PCR or microarrays)
identify groups of SNPS that are linked to disease phenotype
identify nearby genes as candidates for further studies
Genome wide association studeies (GWAS)
used to detect SNP related diseases in large populations
uses large numbers of affected and unaffected individuals
benefits for families too small in size for studies or rare/complex diseases, autosomal recessive mutations
what are the steps in GWAS
identify affected pts and unaffected pts (control)
isolate DNA
analyze SNPs
identify SNP that are frequently present in affected pt
locate the said SNP
identify nearby genes as candidates for further studies
isolation of pt’s DNA
blood
cheek swab
biopsy
saliva
microarrays
steps:
pt sample is isolated, fragmented, and labeled
denature sample→ single stranded
microarray - allow for hybridization (forming double stranded) - only perfect matches can bond to form double stranded DNA
detect flourescence spots (each flourescence spot is an allele A/T/C/G)
compare the pattern w a healthy pt to find variance
application of microarrays
for linkage studies and GWAS (DNA test sample)
for mutation analysis and genotyping (DNA test sample)
gene expression analysis (cDNA is test sample)
study of candidate genes
may be involved in disease process
test for mutations (PCR, microarray, sequencing)
mutations should be present in pt DNA and absent in control
functional analysis of the mutation
develop animal models of disease (transgenic)
develop diagnostic tests for detecting mutaiton
treatments: gene therapy (cloning, crispr)
cloning DNA
allows isolation and manipulation of specific pieces of FNA in lab
determine gene structure
functional analysis
mutational analysis (test for function of mutated genes)
use as probes so southern northern
generate transgenic animals for in vivo studies
develop gene therapies
cloning DNA and cDNA
DNA goes thru PCR to amplify specific target sequence, cut w restriction enzymes to make library containing entire genome
RNA goes thru RT-PCR to amplify single target gene (cDNA), use RT and direct cloning to make library containing enitre transcriptome
cloning vector (plasmid or virus)
bacteria to replicate plasmid
steps in cloning
cut plasmid (circular DNA that replicate in abcteria, carry antibiotic resistance genes to select bacteria)
nsert DNA w restriction enzyme
match, anneal, ligate DNA
feed the ligated DNA to a bacteria (E. coli)
grow bacterial containing plasmid on agar plates
antibiotics in agar prevent growth of Ecoli that do not contain plasmid (due to the antibiotic resistance gene in the plasmid)
isolate plasmid DNA for future uses
mRNA → cDNA
steps:
isolate RNA
RT make double stranded cDNA:
uses Oligo-dT primer binds to poly A tail of mRNA → generate full length cDNA starting 3’ end
PCR
Clone PCR product
Gene therapy for RPE65
clone the therapeutic gene (RT-CPR to generate cDNA and promoter
gene delivery:
adeno-associated virus (AAV) is used
virus is infectious → can infect RPE
virus is replication defective: cannot replicate after infection
AVV produces, stable, long-term expression
treatment: subretinal injection to place virus next to RPE
expression: cells incorporate therapeutic gene and now can make correct RPE65 protein and restore function
transgenic technology
GMO - organisms that has been engineered to contain goreign or modifeied DNA in genome
Random insertion: foreign DNA randomly inserted
KNock-out: deletion of a specific target gene
kock-in: replacement of specfici target w foreign or modified gene
random insertion
clone DNA
inject DNA into nucleus of single cell mouse embryo
DNA inserts into genome in repeated arrays
implant embro into host female
breed pups to analyze phenotypes
pros: fast, can generate many animals in weeks
normal gene still intact
can introduce foreign genes
cons: insertion location random
normal gene still intact
may disrupt unknown genes
insertion sites may alter expression of transgene
targeted gene modif by homologous recombination
goal: replace, delete or modify exisiting gene
replace gene w the desired DNA sequence using crossing over during cell division
applications:
make animal models of disease
study gene function and consequences of specific mutations
generate modif in stem cells for therapies
pros and cons of gene modif by homologous recombination
pros:
can replace or modify specific gene in known location
elimiates off target effects
can be condition to allow gene deletion/alteration only in subset of cells or in response to drug
cons:
difficult, expensive, and time consuming
must be done in dividing cells
methods for gene modif by homologous recombination
isolate embryonic stem cells (ES cells)
clone targeting construct contain desired DNA and antibiotic resistance gene
add targeting construct DNA to ES cells
cells take up DNA and use homologous recombination during DNA rep tro replace target gene w engineered sequence
drug selection to kill any ES cells without new gene
screen resistant colonies for recombinants (vs random)
inject modified ES cells into multiple blastocyst from normal mouse
blastocyst transolanted into female host mouse
resulting progeny are chimeras (mixture of cells w normal and modif genes)
breed w normal mice and genotype pups using PCR to identify pups carrying modified genome
CRISPR-Cas9
clustered regularly interspaced short palndromic repeats
developed from bacterial system used to protect against viral infections
designed to be specific for target gene by virtue of a specific guide DNA sequence
different strategies used to generate deletions (function loss) or gene edits, or to regulate gene expression
use in: any species, cell type, dividing or non-dividing cells
components of CRISPR
Cas9 - bacterial enzyme makes double stranded cuts in DNA at specific location
Guide RNA (gRNA): single stranded RNA molecule containing predesigned guide RNA sequence complementary to targeted hose sequence and tracrRNA scaffold that interacts w Cas9
donor DNA: required when gene edits, used as template to repair double stranded break in host cell DNA
delivery system/vector: for cloning CRISPR/Cas components and delivering to the target cells, uses plasmid or virus containing cDNA Cas9 gene and template for transcribing guide RNA, donor DNA may be separate or engineerred as part of the same construct
method of CRISPR
introduce into cells
guide RNA binds to the target site in host DNA genome
gRNA positions Cas9 at the target site
Cas9 makes a double stranded cut in the host DNA
the cell initiates DNA repair:
without donor DNA: NHEJ generates deletion
with donor DNA: HDR replaces missing sequence using donor sequence